A subscription to JoVE is required to view this content. Sign in or start your free trial.
Live-Cell Imaging of Drosophila melanogaster Third Instar Larval Brains
In This Article
Summary
Here, we discuss a workflow to prepare, dissect, mount, and image live explant brains from Drosophila melanogaster third instar larvae to observe the cellular and subcellular dynamics under physiological conditions.
Abstract
Drosophila neural stem cells (neuroblasts, NBs hereafter) undergo asymmetric divisions, regenerating the self-renewing neuroblast, while also forming a differentiating ganglion mother cell (GMC), which will undergo one additional division to give rise to two neurons or glia. Studies in NBs have uncovered the molecular mechanisms underlying cell polarity, spindle orientation, neural stem cell self-renewal, and differentiation. These asymmetric cell divisions are readily observable via live-cell imaging, making larval NBs ideally suited for investigating the spatiotemporal dynamics of asymmetric cell division in living tissue. When properly dissected and imaged in nutrient-supplemented medium, NBs in explant brains robustly divide for 12-20 h. Previously described methods are technically difficult and may be challenging to those new to the field. Here, a protocol is described for the preparation, dissection, mounting, and imaging of live third-instar larval brain explants using fat body supplements. Potential problems are also discussed, and examples are provided for how this technique can be used.
Introduction
Asymmetric cell division (ACD) is the process by which subcellular components such as RNA, proteins, and organelles are partitioned unequally between daughter cells1,2. This process is commonly seen in stem cells, which undergo ACD to give rise to daughter cells with different developmental fates. Drosophila NBs divide asymmetrically to produce one NB, which retains its stemness, and one ganglion mother cell (GMC). The GMC undergoes further divisions to produce differentiating neurons or glia3. Asymmetrically dividing NBs are abundant in the developing brains of third-instar larvae, which are readily observed via microscopy. At the third instar larval stage, there are roughly 100 NBs present in each central brain lobe3,4,5,6.
Asymmetric cell division is a highly dynamic process. Live-cell imaging protocols have been used to measure and quantify the dynamics of cell polarity7,8,9,10, spindle orientation11,12,13, the dynamics of the actomyosin cortex14,15,16,17,18, microtubule and centrosome biology19,20,21,22,23,24,25,26,27, and membrane10,28 and chromatin dynamics29. Qualitative and quantitative descriptions of ACD rely on robust methods and protocols to image dividing NBs in intact living brains. The following protocol outlines methods to prepare, dissect, and image third instar larval brains for live-cell imaging in vivo using two different mounting approaches. These methods are best suited for researchers interested in the spatiotemporal dynamics of stem cell divisions, as well as divisions in other brain cells, as they allow for short- and long-term observations of cellular events. Additionally, these techniques are readily accessible to newcomers to the field. We demonstrate the effectiveness and adaptability of this approach with larval brains expressing fluorescently tagged microtubule and cortical fusion proteins. We additionally discuss methods of analysis and considerations for application in other studies.
Protocol
NOTE: Figure 1 shows the materials required to perform this study.
1. Considerations and preparations for the experiment
- Prevent the larvae from overcrowding.
NOTE: The quality of explant larval brains is directly related to the health and quality of the larvae prior to dissection. Larvae that are malnourished from overcrowding will generally yield lower-quality brains30.- Ensure that no more than 20-30 larvae are present per meal cap dish to avoid malnutrition. Examples of these can be seen in Figure 2.
- Filter and aliquot Schneider's medium before use.
- For each dissection, prepare fresh imaging and dissection medium by supplementing aliquoted Schneider's insect medium with 1% bovine growth serum (BGS). A volume of 5 mL of dissection and imaging medium is usually sufficient for an imaging experiment.
- Warm the supplemented medium to room temperature (RT) before use.
- Consider the length of the movie to be collected, and use that to factor the supplementation of the imaging medium, the mounting approach, and the acquisition settings of the microscope.
NOTE: Under optimal conditions, NBs in brains supplemented with only BGS will robustly divide for upward of 3 h.- Supplement the imaging medium by adding larval fat body tissues to the imaging medium to support divisions past 4 h when conducting experiments that require longer movies.
NOTE: Fat bodies secrete mitogens that support NB proliferation31, and whole fat bodies from 10 larvae are sufficient to support four to five brains. Further, samples imaged with a membrane-bound slide have been shown to divide for over 10 h13,32, while samples imaged with a multi-well slide usually divide less often (unpublished observations). - Alternatively, implement a more complex imaging medium for longer movies, as described previously33. Minimize photodamage by adjusting the exposure time, laser power, and sampling frequency for best results.
- Supplement the imaging medium by adding larval fat body tissues to the imaging medium to support divisions past 4 h when conducting experiments that require longer movies.
2. Larvae staging and collection (Figure 2)
- Cross 1-5 day old female virgin flies with 1-7 day old adult male flies to produce progeny with the desired genotype. For optimal yield, cross 10-15 female virgins with 5-10 males. Deposit these flies into a fly cage with a meal cap (Figure 2A-C), and incubate at 25 °C.
- Swap the meal cap daily. This prevents the meal caps from becoming overcrowded with larvae, which reduces the quality of the dissected brains.
- If the meal cap is significantly covered in larvae (i.e., >30), split this meal cap in half, and replace one half with a fresh meal cap that has also been cut in half. Alternatively, swap the meal caps on a more frequent basis (i.e., every 12 h instead of every 24 h). Examples of overpopulated meal caps can be seen in Figure 2E, F.
- Incubate the meal cap with larvae at 25 °C until the larvae reach the desired age.
3. Larval fat body dissection (Figure 3)
NOTE: This protocol describes dissections using a 3-well dissection dish.
- Pipette ~400 µL of dissection and imaging medium into each well of a 3-well dissection dish.
- Wash ten 72-96-h old well-fed wild-type larvae by gently holding them with dissection forceps and dipping them in and out of dissection solution in the bottom-most well until all food particulates have been washed off. After rinsing, move the clean larvae to the middle well.
- Using one set of tweezers, hold the larva by the mouth hooks. With the other set of tweezers, rupture one side of the larva's cuticle.
- This rupture will cause the fat bodies to spill out of the larva. The fat bodies are off-white and semi-translucent and will have a lattice-like structure (Figure 3I). The fat bodies will also tend to stick to themselves and the dissection tweezers. Once identified, collect as much fat body from each larva as possible, and transfer it with the forceps to the top-most well with 400 µL of RT dissection medium.
4. Larval brain dissection (Figure 3)
- Wash the experimental larvae in dissection and imaging medium as above to free them of food residues. For best results, avoid storing undissected larvae in the dissection solution. This will cause the larvae to "drown" and will negatively impact the quality of the dissected brains.
- Using one set of tweezers, hold the larva by the mouth hooks. Using another set of tweezers, gently cut/rip off approximately 1/3 of the larva from the posterior side (Figure 3A). This will cause elements of the digestive tract, fat bodies, connective tissue, and nervous system to "burst" out of the ruptured side of the larva (Figure 3B).
- Using one set of tweezers, hold the larva by the mouth hooks. With the other set of tweezers, gently brush the cuticle toward the mouth hooks while "pushing" inward with the tweezers holding the mouth hooks until the entire larva is turned inside out. This motion is similar to turning a sock "inside out" (Figure 3C, D).
- Invert the larva so that the central nervous system and other tissues face outward while still being connected to the cuticle. At this step, locate the central nervous system (CNS) to avoid accidental removal. Using tweezers, gently remove all the non-CNS tissue, leaving only the CNS and brain attached to the cuticle (Figure 3E).
- The brain will be attached to the cuticle via axonal connections. Using microdissection scissors, cut these axonal connections to release the brain from the cuticle. To do this, first gently cut underneath the brain lobes (Figure 3F). Repeat with the connections under the ventral nerve cord.
NOTE: This step may be done with tweezers if microdissection scissors are not available. Take special care when using tweezers to ensure that the brain tissue is not over-stretched during removal from the cuticle because mechanical stress will negatively affect the brain health. - Transfer the dissected brain into a well with dissection and imaging medium. For imaging experiments longer than 3 h, use dissection and imaging medium supplemented with fat bodies as described above. Dissect the larvae in batches to keep the dissection time under 20 min.
5. Mounting and imaging (Figure 4)
- For imaging with a membrane-bound slide34:
- Collect both dissected brains and isolated fat bodies in the last well of the dissection dish.
- Assemble half of the slide by placing a gas-permeable membrane over the back of the slide, and press the split ring into the center, holding it in place (Figure 4A-C).
- Using a 200 µL micropipette, transfer up to 10 dissected brains and as much fat body as possible (see above) in ~130-140 µL of dissection and imaging medium to the membrane. Make sure to deposit the medium with the samples in the center of the gas-permeable membrane (Figure 4D, E).
- Orient the brains for the population of NBs to be imaged and for the type of microscope being used (Figure 4E). Position the sample as close to the microscope's objective as possible. For example, to image NBs in the central brain lobes, orient the brains such that the brain lobes are closest to the objective (Figure 4H).
- Once the brains are oriented, gently place a glass coverslip on top of the solution on the membrane. This will cause the solution containing the brains and fat bodies to spread over the entirety of the membrane (Figure 4F).
- Blot excessive solution by holding a laboratory tissue close to the coverslip edge. The optimal amount of solution is achieved when the brains touch the coverslip without being squashed. If reorientation is required at this step, carefully move the coverslip to move the brains.
- Immobilize the coverslip by applying melted petroleum jelly along the edges of the coverslip with a paintbrush. Allow the jelly to solidify (Figure 4G).
- For imaging with a multi-well imaging slide (Figure 4):
- Add 400 µL of imaging medium to a well of a multi-well slide (in the experiment performed here, a chambered 8-well micro[µ]-slide was used; Figure 4I). Transfer the previously dissected fat bodies to this well (see step 3.4).
- Deposit up to 10 brains in a cluster near the center of the well (Figure 4J).
- Orient the brains for the population of NBs to be imaged and for the type of microscope being used, as described in step 5.1.4 (Figure 4K). Arrange the samples so they are close to each other. This will minimize the distance the stage must move between samples, which reduces sample drift during acquisition.
- Once the brains have been oriented in the well, allow the brains to settle for 2-5 min. This increases their stability during transport/imaging. Prepare the microscope for acquisition during this time.
- Cover the µ-slide with the slide cover, and transfer it to the microscope. Begin acquisition with the lowest laser power and exposure time possible to minimize photobleaching.
6. Data processing and management best practices
- Process the data as needed according to the available analysis software.
- For the example shown here, save the acquired data with SlideBook software as a SlideBook Image File (.sld).
- To convert into Imaris' proprietary file type (.ims) using the Imaris File Converter, open the Imaris File Converter in a separate window. Click on and drag the .sld files into the " input" section of the Imaris File Converter.
- Determine the desired output location for the converted files, and click on "Start all."
- After conversion, view and annotate the data in the Imaris software.
NOTE: Alternatives for image analysis can be used in place of Imaris, such as Fiji (https://hpc.nih.gov/apps/Fiji.html), Aivia (https://www.aivia-software.com/), Volocity (https://www.volocity4d.com/), or others.
- Retain as much of the original data as possible for proper record-keeping. For example, if the acquisition software is saved in one file format but is converted to a different format for analysis, retain the acquired version of the data.
- For data analysis, maintain a record of as many details as possible about the sample and acquisition settings. Key information to retain includes the genotype of the dissected larvae, the age of the larvae prior to dissection, the state of the meal cap they were reared in, the laser power used during imaging, the exposure time, the length of acquisition, and the temporal resolution.
7. Example quantification of cell cycle length (Figure 5)
NOTE: in this example, larvae expressing the polarity marker Pins (Pins::EGFP16) and the microtubule-binding protein Jupiter25 (cherry::Jupiter13) were imaged. The subsequent analysis was performed using Imaris software.
- Open the movie using the image analysis software of choice. Scroll through the length of the movie to identify dividing NBs, and label them for future reference. Identify the dividing NBs by their distinct mitotic spindles (Figure 5C-E).
- Identify a reference cell cycle stage to determine the cell cycle length. In this example, metaphase is used as a reference.
- Manually determine the number of frames between successive metaphases, and convert it to minutes or hours to determine the time taken to complete one cell cycle.
- Do this by taking the temporal resolution of the movie and multiplying it by the number of frames between metaphases. For example, if the temporal resolution of the movie is one frame every 5 min, and metaphases are observed in frame 13 and frame 35, the time between these metaphases would be 110 min ([35 − 13] × 5).
- Plot the data with any appropriate software. The data shown here were plotted using PRISM software.
8. Example quantification of cell spindle alignment (Figure 5)
NOTE: In this example, the analysis is performed using Imaris software.
- Open the movie file in Imaris or another software of choice. Scroll through the length of the movie to identify dividing NBs, and label them for future reference.
- Determine the vector formed by the spindle poles using the apical and basal centrosomes (represented by m), as follows:
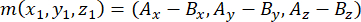
where Ax, Ay, and Az are the coordinates of the apical centrosome, and Bx, By, and Bz are the coordinates of the basal centrosome. Similarly, the axis of the division vector (represented by n) is formed by the midpoint of the apical Pins::EGFP crescent and the basal cortex:
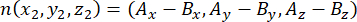
where Ax, Ay, and Az are the coordinates of the midpoint of the Pins::EGFP crescent, and Bx, By, and Bz are the coordinates of the midpoint of the basal cortex. - Determine the magnitude of the vectors m and n:
Magnitude of m: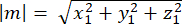
Magnitude of n: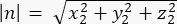
- Determine the dot product (represented by k) of m and n:
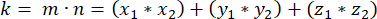
- Using the dot product k and vector magnitudes m and n, determine the angle between the vectors:
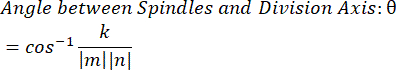
- Plot the data in the software of choice. The data shown here were prepared in Microsoft Excel and visualized in PRISM.
Results
Dissection and imaging of central brain lobe NBs expressing Pins::EGFP and Cherry::Jupiter
To showcase this protocol, larvae expressing UAS-driven Cherry::Jupiter13 and endogenously tagged Pins::EGFP16 (w; worGal4, UAS-cherry::jupiter/CyO; Pins::EGFP/TM6B, Tb) were imaged for 4 h using the described protocol using multi-well imaging slides (Figure 5C,D). Additional data were taken from larvae expressing U...
Discussion
This protocol outlines one approach for the imaging of live explant brains from Drosophila melanogaster larvae. The protocol described here allows for explant brains to be observed for 12-20 h under the right experimental conditions. Special consideration must be given to the preparation of samples and the design of the desired experiments. As mentioned above, one of the most critical factors that determines the quality of the dissected tissue is the health of the larvae. To achieve the highest quality possible,...
Disclosures
The authors have no financial disclosures to declare.
Acknowledgements
This research is supported by R35GM148160 (C. C.) and a National Institutes of Health (NIH) Training Grant T32 GM007270 (R. C. S)
Materials
| Name | Company | Catalog Number | Comments |
| 0.22 µm polyethersulfone (PES) Membrane | Genesee | 25-231 | Vacuum-driven filters |
| Agar | Genesee | 20-248 | granulated agar |
| Analytical Computer | Dell | NA | Intel Xeon Gold 5222 CPU with two 3.80 GHz processors running Windows 10 on a 64-bit operating system |
| Bovine Growth Serum | HyClone | SH30541.02 | |
| Chambered Imaging Slides | Ibidi | 80826 | |
| Confocal Microscope | Nikon | NA | |
| Custom-machined metal slide | NA | NA | See Cabernard and Doe 2013 (Ref. 34) for specifications |
| Dissection Dishes | Fisher Scientific | 5024343 | 3-well porcelain micro spot plate |
| Dissection Forceps | World Precision Instruments | Dumont #5 | |
| Dissection Microscope | Leica | NA | |
| Dissection Scissors | Fine Science Tools (FST) | 15003-08 | |
| Embryo collection cage | Genesee | 59-100 | |
| Flypad with access to CO2 to anesthetize adult flies | Genesee | 59-172 | |
| Gas-permeable membrane | YSI | 98095 | Gas-permeable membrane |
| Glass Cover Slides | Electron Microscopy Sciences | 72204-03 | # 1.5; 22 mm x 40 mm glass coverslips |
| Imaris | Oxford Instruments | NA | Alternatives: Fiji, Volocity, Aivia |
| Imaris File Converter | Oxford Instruments | NA | |
| Instant Yeast | Saf-Instant | NA | |
| Molasses | Genesee | 62-117 | |
| Petri dish | Greiner Bio-One | 628161 | 60 mm x 15 mm Petri dish |
| Petroleum Jelly | Vaseline | NA | |
| Schneider's Insect Medium with L-glutamine and sodium bicarbonate liquid | Millipore Sigma | S0146 | |
| SlideBook acquisition software | 3i | NA | |
| Vacuum-Driven Filtration Unit with a 0.22 µµm PES membrane filter | Genesee Scientific, GenClone | 25-231 |
References
- Delgado, M. K., Cabernard, C. Mechanical regulation of cell size, fate, and behavior during asymmetric cell division. Current Opinion in Cell Biology. 67, 9-16 (2020).
- Sunchu, B., Cabernard, C. Principles and mechanisms of asymmetric cell division. Development. 147 (13), (2020).
- Homem, C. C. F., Knoblich, J. A. Drosophila neuroblasts: A model for stem cell biology. Development. 139 (23), 4297-4310 (2012).
- Gallaud, E., Pham, T., Cabernard, C. Drosophila melanogaster neuroblasts: A model for asymmetric stem cell divisions. Results and Problems in Cell Differentiation. 61 (1489), 183-210 (2017).
- Loyer, N., Januschke, J. Where does asymmetry come from? Illustrating principles of polarity and asymmetry establishment in Drosophila neuroblasts. Current Opinion in Cell Biology. 62, 70-77 (2020).
- Pollington, H. Q., Seroka, A. Q., Doe, C. Q. From temporal patterning to neuronal connectivity in Drosophila type I neuroblast lineages. Seminars in Cell & Developmental Biology. 142, 4-12 (2023).
- Oon, C. H., Prehoda, K. Asymmetric recruitment and actin dependent cortical flows drive the neuroblast polarity cycle. eLife. 8, e45815 (2019).
- Ramat, A., Hannaford, M., Januschke, J. Maintenance of miranda localization in Drosophila neuroblasts involves interaction with the cognate mRNA. Current Biology. 27 (14), 2101-2111 (2017).
- Oon, C. H., Prehoda, K. E. Phases of cortical actomyosin dynamics coupled to the neuroblast polarity cycle. eLife. 10, e66574 (2021).
- LaFoya, B., Prehoda, K. E. Actin-dependent membrane polarization reveals the mechanical nature of the neuroblast polarity cycle. Cell Reports. 35 (7), 109146 (2021).
- Siller, K. H., Doe, C. Q. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Developmental Biology. 319 (1), 1-9 (2008).
- Siller, K. H., Cabernard, C., Doe, C. Q. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nature Cell Biology. 8 (6), 594-600 (2006).
- Cabernard, C., Doe, C. Q. Apical/basal spindle orientation is required for neuroblast homeostasis and neuronal differentiation in Drosophila. Developmental Cell. 17 (1), 134-141 (2009).
- Cabernard, C., Prehoda, K. E., Doe, C. Q. A spindle-independent cleavage furrow positioning pathway. Nature. 467 (7311), 91-94 (2010).
- Connell, M., Cabernard, C., Ricketson, D., Doe, C. Q., Prehoda, K. E. Asymmetric cortical extension shifts cleavage furrow position in Drosophila neuroblasts. Molecular Biology of the Cell. 22 (22), 4220-4226 (2011).
- Tsankova, A., Pham, T. T., Garcia, D. S., Otte, F., Cabernard, C. Cell polarity regulates biased myosin activity and dynamics during asymmetric cell division via Drosophila rho kinase and protein kinase N. Developmental Cell. 42 (2), 143-155 (2017).
- Montembault, E., et al. Myosin efflux promotes cell elongation to coordinate chromosome segregation with cell cleavage. Nature Communications. 8 (1), 326 (2017).
- Roubinet, C., et al. Spatio-temporally separated cortical flows and spindle geometry establish physical asymmetry in fly neural stem cells. Nature Communications. 8 (1), 1383 (2017).
- Januschke, J., et al. Centrobin controls mother-daughter centriole asymmetry in Drosophila neuroblasts. Nature Cell Biology. 15 (3), 241-248 (2013).
- Januschke, J., Llamazares, S., Reina, J., Gonzalez, C. Drosophila neuroblasts retain the daughter centrosome. Nature Communications. 2 (1), 243 (2011).
- Rebollo, E., et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Developmental Cell. 12 (3), 467-474 (2007).
- Januschke, J., Gonzalez, C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. The Journal of Cell Biology. 188 (5), 693-706 (2010).
- Rusan, N. M., Peifer, M. A role for a novel centrosome cycle in asymmetric cell division. The Journal of Cell Biology. 177 (1), 13-20 (2007).
- Lerit, D. A., et al. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. Journal of Cell Biology. 210 (1), 79-97 (2015).
- Gallaud, E., et al. Dynamic centriolar localization of Polo and Centrobin in early mitosis primes centrosome asymmetry. PLoS Biology. 18 (8), e3000762 (2020).
- Ramdas Nair, A., et al. The microcephaly-associated protein Wdr62/CG7337 is required to maintain centrosome asymmetry in Drosophila neuroblasts. Cell Reports. 14 (5), 1100-1113 (2016).
- Singh, P., Nair, A. R., Cabernard, C. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Current Biology. 24 (13), 1548-1555 (2014).
- LaFoya, B., Prehoda, K. E. Consumption of a polarized membrane reservoir drives asymmetric membrane expansion during the unequal divisions of neural stem cells. Developmental Cell. 1534 (23), 00159 (2023).
- Sunchu, B., et al. Asymmetric chromatin retention and nuclear envelopes separate chromosomes in fused cells in vivo. Communications Biology. 5 (1), 953 (2022).
- Oliveira, A. C., Rebelo, A. R., Homem, C. C. F. Integrating animal development: How hormones and metabolism regulate developmental transitions and brain formation. Developmental Biology. 475, 256-264 (2021).
- Britton, J. S., Edgar, B. A. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 125 (11), 2149-2158 (1998).
- Lee, C. -. Y., et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes & Development. 20 (24), 3464-3474 (2006).
- Homem, C. C. F., Reichardt, I., Berger, C., Lendl, T., Knoblich, J. A. Long-term live cell imaging and automated 4D analysis of Drosophila neuroblast lineages. PLoS ONE. 8 (11), e79588 (2013).
- Cabernard, C., Doe, C. Q. Live imaging of neuroblast lineages within intact larval brains in Drosophila. Cold Spring Harbor Protocols. 2013 (10), 970-977 (2013).
- Karpova, N., Bobinnec, Y., Fouix, S., Huitorel, P., Debec, A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motility and the Cytoskeleton. 63 (5), 301-312 (2006).
- Loyer, N., Januschke, J. The last-born daughter cell contributes to division orientation of Drosophila larval neuroblasts. Nature Communications. 9 (1), 3745 (2018).
- Bostock, M. P., et al. An immobilization technique for long-term time-lapse imaging of explanted Drosophila tissues. Frontiers in Cell and Developmental Biology. 8, 590094 (2020).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved