A subscription to JoVE is required to view this content. Sign in or start your free trial.
Single-Cell Sorting of Immunophenotyped Mesenchymal Stem Cells from Human Exfoliated Deciduous Teeth
In This Article
Summary
This protocol describes the use of fluorescence-activated cell sorting of human mesenchymal stem cells using the single-cell sorting method. Specifically, the use of single-cell sorting can achieve 99% purity of the immunophenotyped cells from a heterogeneous population when combined with a multiparametric flow cytometry-based approach.
Abstract
The mesenchymal stem cells (MSCs) of an organism possess an extraordinary capacity to differentiate into multiple lineages of adult cells in the body and are known for their immunomodulatory and anti-inflammatory properties. The use of these stem cells is a boon to the field of regenerative biology, but at the same time, a bane to regenerative medicine and therapeutics owing to the multiple cellular ambiguities associated with them. These ambiguities may arise from the diversity in the source of these stem cells and from their in vitro growth conditions, both of which reflect upon their functional heterogeneity.
This warrants methodologies to provide purified, homogeneous populations of MSCs for therapeutic applications. Advances in the field of flow cytometry have enabled the detection of single-cell populations using a multiparametric approach. This protocol outlines a way to identify and purify stem cells from human exfoliated deciduous teeth (SHEDs) through fluorescence-assisted single-cell sorting. Simultaneous expression of surface markers, namely, CD90-fluorescein isothiocyanate (FITC), CD73-peridinin-chlorophyll-protein (PerCP-Cy5.5), CD105-allophycocyanin (APC), and CD44-V450, identified the "bright," positive-expressors of MSCs using multiparametric flow cytometry. However, a significant drop was observed in percentages of quadruple expressors of these positive markers from passage 7 onwards to the later passages.
The immunophenotyped subpopulations were sorted using the single-cell sort mode where only two positive and one negative marker constituted the inclusion criteria. This methodology ensured the cell viability of the sorted populations and maintained cell proliferation post sorting. The downstream application for such sorting can be used to evaluate lineage-specific differentiation for the gated subpopulations. This approach can be applied to other single-cell systems to improve isolation conditions and for acquiring multiple cell surface marker information.
Introduction
Mesenchymal stem cells (MSCs) can be regarded as a scalable source of cells suitable for cell-based therapies and may be considered a gold standard system in regenerative medicine. These cells can be isolated from a variety of sources in the body with different tissue origins1. Depending on their source tissue, each type of MSC displays an ambiguous in vitro behavior2. This is well observed in their morphological and functional properties3. Multiple studies have shown intra-clonal variation in dimensions, including adult tissue differentiation, genomic state, and metabolic and cellular architecture of MSCs2,4.
Immunophenotyping of cells has been a common application of flow cytometry for the identification of stem cells and this was utilized by the International Society for Cell and Gene Therapy (ISCT) in 2006 to prescribe a list of minimal criteria to identify cells as MSCs. It stated that along with plastic adherence and the ability to differentiate into three lineages (osteogenic, chondrogenic, and adipogenic) in vitro, ≥95% of the cell population must express CD105, CD73, CD90, and these cells must lack the expression (≤2% positive) of CD34, CD45, CD11b, CD14, and HLA-DR, as measured by flow cytometry5. Although the MSCs were defined by a set of biomarkers under the minimal criteria of the ISCT, their immune properties could not be benchmarked with these biomarkers and there was a need for more beyond these criteria to make cross-study comparisons and clonal variations easier to quantify2.
Despite the guidelines set by ISCT, extensive research on MSCs has shown that heterogeneity exists in this population, which could arise due to a multitude of factors, mainly due to the ubiquitous nature of heterogeneity that arises between MSC donors6, tissue sources7, individual cells within a clonal population8, and culture conditions2,9,10. Characterization and purification of these primary cells from a variety of tissue sources to ensure quality and cell fate are key steps in their production. The need to understand the displayed variations amongst the population requires an efficient method to resolve it into subpopulations that can be divided and collected separately11. Single-cell level analyses help overcome the challenges of cell-cell variation, reduce biological noise arising from a heterogeneous population, and offer the ability to investigate and characterize rare cells12.
Based on the purpose and chosen parameters, several methods can be employed to sort and enrich the selected populations. Cell-sorting techniques can comprise both bulk sorting and single-cell sorting methods. While bulk sorting can enrich target populations through Magnetic-activated cell sorting (MACS)13, fractionation14, and elutriation15, single-cell sorting can enrich more homogeneous populations by means of fluorescence-activated cell sorting (FACS)11. A comparative analyses of each of these methods with its own set of advantages and disadvantages is highlighted in Table 1.
Table 1: Comparative analyses of different techniques: MACS, Fractionation, Elutriation, and FACS highlighting the differences in their principle and the advantages and disadvantages of choosing a particular technique over another. Abbreviations: MACS = Magnetic-activated cell sorting; FACS = Fluorescence-activated cell sorting. Please click here to download this Table.
Since the advent of the technique, single-cell flow cytometry has played a major role in enumeration16, detection, and characterization of a specific cell population in a heterogeneous sample17. Hewitt et al. in 2006 laid the foundation of automated cell sorting methodology to enhance the isolation of homogenous pools of differentiated human embryonic stem cells (hESCs)18. Single-cell sorting enriched the population of GFP-transduced hESCs facilitating the isolation of genetically modified clones, which opened a new dimension in clinical research. To improve the sort efficiency, two approaches have generally been taken; either the collection media of the sorted populations are modified to sustain viability and proliferation of post-sorted cells19 or the cell-sorting algorithm/software is appropriately modified12.
With the advancement of technology, commercial flow cytometers and cell sorters have been able to help address challenges that were met while aseptically sorting fragile and rare cell populations, especially stem cells of different origins. One of the major challenges of stem cell biologists has been the clonal isolation of human pluripotent stem cells following transfection protocols required in gene editing studies19. This was addressed by sorting single cells into 96-well plates that were coated with Mouse Embryonic Fibroblasts (MEFs) along with supplements and commercial small molecule ROCK inhibitors. However, cell isolation strategies could be largely refined with the use of index sorting, a feature of the sorting algorithm that identifies the immunophenotype of individual cells sorted12. This refined modality in single-cell sorting helped not only in enhancing sort efficiency for stem cells, especially with regard to rare hematopoietic stem cell populations, but also efficiently linked single-cell clones to their downstream functional assays20.
This paper focuses on single-cell sorting of immunophenotyped stem cells from human exfoliated deciduous teeth (SHEDs) for the enrichment of sub-populations to study their functional differentiation capacities. Using a combination of two MSC-positive markers, CD90 and CD73, and a negative hematopoietic marker CD45, the MSCs were immunophenotyped and the dim and null expressors were identified. Based on their immunophenotype the subpopulations were identified as pure MSCs, single positive and double negative populations. They were sorted using the single-cell sort mode to obtain pure and enriched subpopulations for further functional studies to identify whether the differential expression of markers was an artifact of in vitro culture conditions or whether it has any effect on the functional properties as well. Cells that were not homogeneous expressors of the "positive MSC markers" were sorted to study their functional properties.
Protocol
Ethics approval and consent to participate: Human exfoliated deciduous dental pulp samples were received after obtaining informed consent and full ethical approval by Sri Rajiv Gandhi Dental College and Hospital (SRGCDS) Oral and Maxillofacial Department, Bengaluru, in accordance with the standards established by the Hospital Ethical Clearance Committee, SRGCDS. Following which isolation, culture, maintenance, and application of SHEDs were approved by and in compliance with the guidelines recommended by the Institutional Committee for Stem Cell Research (IC-SCR) at Manipal Institute of Regenerative Medicine, MAHE - Bengaluru. See the Table of Materials for details about all materials and reagents used in this protocol.
1. Preparation of reagents and buffers
- For culture maintenance
- Prepare MSC cell culture media (10%) using basal medium for undifferentiated hESCs, 10% fetal bovine serum (FBS), 1% L-glutamine, and 1% penicillin-streptomycin (Pen-strep) (Table 2).
- Prepare MSC cell culture media (20%) using basal medium for undifferentiated hESCs, 20% FBS, 1% L-glutamine, and 1% Pen-strep (Table 2).
- Prepare neutralizing media using basal medium for undifferentiated hESCs, 1% L-glutamine, and 1% antibiotic-antimycotic (Anti-Anti) (Table 2).
- For flow cytometric analysis and sorting
- Prepare staining buffer using 2% FBS in phosphate-buffered saline (PBS).
- Prepare a stock solution of 4′,6-diamidino-2-phenylindole (DAPI) (1 µg/mL) by adding 1 µL in 1 mL of PBS
- For lineage-specific differentiation of MSCs
- Prepare differentiation media for osteogenic, chondrogenic, and adipogenic differentiation according to the composition described in Table 3. Store the prepared aliquots at 4 °C for the duration of the experiment.
- Prepare serum-starved media (2% media) using basal medium for undifferentiated hESCs, 2% FBS, 1% L-glutamine, and 1% Pen-strep (Table 2).
| TYPE OF MEDIA | PURPOSE OF MEDIA | COMPOSITION FOR 50 mL | ||||||||
| FBS | Pen-Strep | L-Glutamine | BASAL MEDIUM FOR UNDIFFERENTIATED hESCs | |||||||
| 10% media | MSC culture and maintenance | 5 mL | 500 μL | 500 μL | 44 mL | |||||
| 20% media | CFU-F assay | 10 mL | 500 μL | 500 μL | 39 mL | |||||
| Serum starved (2%) media | Media for Control wells in Differentiation protocol | 1 mL | 500 μL | 500 μL | 48 mL | |||||
| Neutralizing media | Media for neutralizing the cell suspension after trypsinization | - | 500 μL | 500 μL | 49 mL | |||||
Table 2: Cell culture media for culture maintenance and assays. Abbreviations: MSC = mesenchymal stem cell; CFU-F = colony-forming unit-fibroblast.
| COMPONENTS | OSTEOGENIC MEDIA | CHONDROGENIC MEDIA | ADIPOGENIC MEDIA |
| Basal Media | 90 mL | 90 mL | 90 mL |
| Induction media | 10 mL | 10 mL | 10 mL |
| Total Volume | 100 mL | 100 mL | 100 mL |
Table 3: Differentiation media for trilineage differentiation of SHEDs.
2. Culture and maintenance of SHEDs
- Maintain cells in 10% MSC culture media and perform media changes every 2 days or as required.
- Trypsinize cells at 95% confluency using 0.25% trypsin-EDTA.
- Neutralize cells after trypsinization using neutralizing media.
- Centrifuge the tube at 300 × g for 6 min at room temperature to obtain a cell pellet.
- Decant the supernatant and resuspend the cell pellet in 10% MSC culture media.
- Seed cells into freshly prepared cell culture dishes containing 10% MSC culture media for further experiments or sub-culturing.
NOTE: Optimal seeding density for SHEDs is 0.2 × 106 cells in a 100 mm dish and 0.8 × 106 cells in a T-75 flask, to obtain 1.5 × 106 cells and 4 × 106 cells, respectively, at 90-100% confluency.
3. Characterization of MSCs
- Short-term cell growth assay
- Seed 4 × 104 cells/well in triplicate into 6-well plates in 10% MSC culture media.
- Incubate the plates for 7 days at 37 °C, and perform media changes every 2 days.
- Harvest the cells on days 2, 4, and 8 using 0.25% trypsin treatment and wash with culture media.
- Centrifuge the cells at 300 g for 6 min at room temperature and resuspend the pellet in 1 mL of media.
- Count the cells using a hemocytometer and determine their viability by the trypan blue exclusion method.
- Calculate the proliferation rate using equation (1):
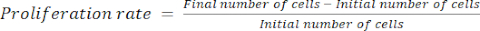 (1)
(1)
- Colony-forming unit-fibroblast (CFU-F) assay
- Seed 10,000 cells in a 100 mm dish and culture them in 20% MSC culture media.
- Incubate the plates for 14 days at 37 °C, and perform media changes every 3 days.
- After 14 days, rinse the colonies with PBS, fix them with crystal violet dye in methanol, and rinse again with PBS to remove the residual stain.
- Count and image the colonies.
NOTE: Count colonies with >50 cells. Colony-forming efficiency is calculated as colony-forming unit numbers.
- Immunofluorescence assay
- Seed the MSCs on 35 mm dishes and let them grow till 80-90% confluency.
- Remove the media and rinse the dishes with PBS once.
- Fix the cells with 1 mL of 4% paraformaldehyde (PFA) by either incubating for 1 h at room temperature or overnight at 4 °C.
- After fixation, wash the wells with PBST for 3 x 5 min on the rocker.
- Add 0.3% Triton X-100 in PBST (0.05% Tween 20 solution in PBS) for permeabilizing the cells. Keep it on the rocker for 15 min at room temperature.
- Wash the wells with PBST for 3 x 5 min on the rocker.
- Add 3% bovine serum albumin (BSA) for blocking and keep it on the rocker for 1 h at room temperature.
- Wash the wells with PBST for 3 x 5 min on the rocker.
- Add 800 µL of 1:500 dilution of mouse anti-vimentin, keep it on the rocker for 1 h at room temperature, and transfer the plate to 4 °C for overnight incubation.
- Next day, remove the primary antibody and wash the wells with PBST for 3 x 5 min on the rocker.
- Add 800 µL of 1:1,000 dilution of goat anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 555, and keep it for 3 h at room temperature on the rocker.
- Wash the wells with PBST for 3 x 5 min on the rocker.
- Add Alexa fluor 488 Phalloidin Probes 240 µL in 1,000 µL of PBS and incubate at room temperature for 60 min on the rocker.
- Wash the wells with PBST for 3 x 5 min on the rocker.
- Add 700 µL of DAPI mountant and observe the cells under a microscope.
- Lineage-specific differentiation
- Differentiation following 2D culture
- Take two 48-well plates and label them as osteogenic and adipogenic lineages, respectively.
- Seed 15,000 cells/well in four wells of each plate and culture them in 10% MSC culture media.
- Once the monolayer of cells has reached 90% confluency, label the first two wells as 'Control' and replace the existing media with serum-starved media (2% media). In the last two wells labeled as 'Test,' add differentiation media of either of the two lineages, adipogenic or osteogenic. Mark this as Day 0.
- Gently replace media every third day to circumvent peeling and be careful to avoid contamination.
- Maintain these conditions till the twenty-first day; then process for the staining experiment.
- Differentiation following 3D culture
- Take two 15 mL tubes for performing chondrogenic differentiation using 3D pellet culture.
- Transfer 1 × 106 cells to each tube and centrifuge at 300 × g for 6 min to form a pellet. Label one tube as 'Control' and add 10% MSC culture media to it; label the other tube as 'Test' and add chondrogenic differentiation media. Place the tubes carefully in the incubator with their caps loosely screwed. Mark this as Day 0.
- Change the media every third day carefully, so as not to dislodge/disintegrate the pellet during media changes.
- Maintain these conditions till the twenty-first day and then, process the cells for further experiments.
- Differentiation following 2D culture
- Lineage-specific cytochemical staining
- For 2D cultures, use the differentiated (test) and undifferentiated MSCs (control) plates (from step 3.4.1.5.) for staining, and first, remove the media and wash twice with PBS. Fix the cells using 4% PFA for 30 min at room temperature, remove the supernatant, and wash once with PBS. Perform staining for each lineage as follows.
- For adipocyte lineage, add permeabilization solution from the kit and incubate the plate for 5 min at room temperature. Prepare and add 1 mL of Oil red O working solution and keep it for 10 min. Remove the stain and give five washes with distilled water.
- For osteocyte lineage, add 5% of freshly prepared silver nitrate (in distilled water) to each well and keep the plate under UV for 1 h. Remove the solution and add 2.5% of sodium thiosulphate to remove the unreacted silver; keep it for 5 min. Remove the stain, wash twice with distilled water, and observe the stained cells under a microscope.
- For 3D cultures, collect the pellet after the end of the differentiation period from step 3.4.2.3 and obtain cryosections of the differentiated tissue in the form of the pellet. Allow the slides to air dry and be at room temperature before proceeding to the staining.
- For chondrocyte lineage, follow the directions of the staining kit to add a sufficient volume of washing solution, remove it, add the fixing solution, and incubate for 30 min. Wash with distilled water, add the staining solution, and incubate for 30 min. Wash thrice with 0.1 N hydrochloric acid; add distilled water to neutralize the acidity. Observe the stained cells under a brightfield microscope.
- For 2D cultures, use the differentiated (test) and undifferentiated MSCs (control) plates (from step 3.4.1.5.) for staining, and first, remove the media and wash twice with PBS. Fix the cells using 4% PFA for 30 min at room temperature, remove the supernatant, and wash once with PBS. Perform staining for each lineage as follows.
4. Cell surface staining for immunophenotyping
NOTE: Recommended cell culture plates for getting an optimal number of cells in steps 4.2-4.5 are 100 mm dishes or T75 flasks.
- Cell preparation for flow cytometric experiments
- Trypsinize and collect the cells from a confluent dish/ flask and centrifuge at 300 × g for 6 min to obtain the cell pellet.
- Resuspend the pellet in 1 mL of media and determine the viable cell count using a hemocytometer following the trypan blue exclusion method.
- Centrifuge the cell suspension again after counting and give two more washes to the pellet with 1 mL of staining buffer.
- Discard the supernatant and finally resuspend the pellet in an appropriate volume of the staining buffer depending on the protocol (see steps 4.2 to 4.5).
- Preparation of compensation controls
- Take seven FACS tubes and label them as Unstained, DAPI, V450, FITC, PE, PerCP-Cy 5.5, and APC.
- Resuspend the final pellet in the staining buffer (see step 4.1.) keeping a cell density of 0.5 × 106 cells per 50 µL per tube for the Unstained and DAPI tubes.
- Prepare the single-stained tubes for compensation as described in Table 4.
- Gently vortex each tube after preparation and incubate in the dark for 30 min.
- After incubation, give two washes by adding 1 mL of staining buffer to each tube, followed by brief vortexing and centrifugation at 200 g for 10 min at room temperature.
- Discard the supernatant, resuspend the pellet in 500 µL of staining buffer, and set it aside until the acquisition.
- For the DAPI tube, perform heat shock treatment by incubating it in a 60 °C water bath for 5 min followed by 15 min on ice. Add 5 µL of DAPI to the suspension and keep it in the dark until the run acquisition.
- Preparation of fluorescence minus one (FMO) controls
- Resuspend the final pellet in staining buffer (see step 4.1) keeping a cell density of 0.5 × 106 cells per 50 µL for each tube.
- Take five FACS tubes, and label them as CD44-V450 isotype, CD90-FITC, CD45-PE, CD73-PerCP Cy5.5 isotype, and CD105-APC isotype.
- Prepare the cell and antibody suspension according to Table 5.
- Vortex the tubes gently and incubate them for 30 min at room temperature in the dark.
- After incubation, give two washes with 1 mL of staining buffer to each tube followed by brief vortexing and centrifugation at 200 g for 10 min at room temperature.
- Discard the supernatant, resuspend the pellet in 500 µL of staining buffer, and set it aside until acquisition.
- Preparation of samples for analysis
- Resuspend the final pellet in the staining buffer (see step 4.1) keeping a cell density of 0.5 × 106 cells per 50 µL for each tube.
- Take two FACS tubes and label them as Mixed tubes 1 and 2.
- Prepare the cell and antibody suspension according to Table 6.
- Vortex the tubes gently and incubate them for 30 min at room temperature in the dark.
- After incubation, give two washes with 1 mL of staining buffer to each tube followed by brief vortexing and centrifugation at 200 g for 10 min at room temperature.
- Discard the supernatant, resuspend the pellet in 500 µL of staining buffer, and set aside until acquisition.
- Preparation of samples for single-cell sorting
- Take two FACS tubes, add 1mL of FBS, and roll the tube around till an even layer of FBS is formed on the inside of each tube. Incubate this for 1-2 h while processing the sample for staining. Label these tubes as Mixed tubes 1 and 2.
- Resuspend the final pellet in the staining buffer (see step 4.1) keeping a cell density of 2-3 × 106 cells per 50 µL for each tube.
- Prepare the cell and antibody suspension in Mixed tubes 1 and 2 according to Table 7.
- Vortex the tubes gently and incubate them for 30 min at room temperature in the dark.
- After incubation, give two washes with 1 mL of staining buffer to each tube followed by brief vortexing and centrifugation at 200 g for 10 min at room temperature.
- Discard the supernatant, resuspend the pellet in 500 µL of staining buffer, and set aside. Add 5 µL of DAPI 15 min prior to sorting.
NOTE: Note that for sorting experiments higher cell density is recommended per tube.
| Tube type | Positive Comp Beads* | Negative Comp Beads* | Cells | Antibody added | |
| FITC tube | 1 drop | 1 drop | – | Anti-human CD90-FITC (2 µL) | |
| V450 tube | 1 drop | 1 drop | – | Anti-human CD44-V450 (2 µL) | |
| PerCP-Cy 5.5 tube | 1 drop | 1 drop | – | Anti-human CD73-PerCP Cy 5.5 (2 µL) | |
| PE tube | 1 drop | 1 drop | – | Anti-human CD45-PE (2 µL) | |
| APC tube | 1 drop | 1 drop | – | Anti-human CD105-APC (2 µL) | |
| DAPI tube | – | – | 50 µL | – | |
| Unstained tube | – | – | 50 µL | – | |
| *1 drop= 60 µL of bead suspension | |||||
Table 4: Compensation control samples. Abbreviations: Comp = compensation; DAPI = 4',6-diamidino-2-phenylindole; FITC = fluorescein isothiocyanate; APC = allophycocyanin; PE = phycoerythrin; PerCP = peridinin-chlorophyll-protein.
| TUBE TYPE | ANTIBODY AGAINST POSITIVE MARKER (2 µL) | ANTIBODY AGAINST NEGATIVE MARKER (2 µL) | ISOTYPE ANTIBODIES ADDED (2 µL) | TOTAL VOLUME OF ANTIBODIES | VOLUME OF CELL SUSPENSION ADDED | VOLUME OF STAINING BUFFER ADDED | ||
| CD90-FITC FMO tube | - Anti-human CD44-V450 | Anti-human CD45-PE | FITC IgG1 isotype | 10 µL | 50 µL | 40 µL | ||
| - Anti-human CD73 PerCP Cy 5.5 | ||||||||
| - Anti-human CD105-APC | ||||||||
| CD73-PerCP Cy5.5 FMO tube | - Anti-human CD44-V450 | Anti-human CD45-PE | PerCP Cy 5.5 IgG1 isotype | 10 µL | 50 µL | 40 µL | ||
| - Anti-human CD90-FITC | ||||||||
| - Anti-human CD105-APC | ||||||||
| CD44-V450 FMO tube | - Anti-human CD90-FITC | Anti-human CD45-PE | V450 IgG1 isotype | 10 µL | 50 µL | 40 µL | ||
| - Anti-human CD73-PerCP Cy 5.5 | ||||||||
| - Anti-human CD105-APC | ||||||||
| CD105-APC FMO tube | - Anti-human CD44-V450 | Anti-human CD45-PE | APC IgG1 isotype | 10 µL | 50 µL | 40 µL | ||
| - Anti-human CD90-FITC | ||||||||
| - Anti-human CD73-PerCP Cy 5.5 | ||||||||
| CD45-PE FMO tube | - Anti-human CD44-V450 | - | PE IgG1 isotype | 10 µL | 50 µL | 40 µL | ||
| - Anti-human CD90-FITC | ||||||||
| - Anti-human CD73-PerCP Cy 5.5 | ||||||||
| - Anti-human CD105-APC | ||||||||
Table 5: FMO control samples. Abbreviations: FMO = fluorescence minus one; FITC = fluorescein isothiocyanate; APC = allophycocyanin; PE = phycoerythrin; PerCP = peridinin-chlorophyll-protein.
| TUBE TYPE | ANTIBODY AGAINST POSITIVE MARKER (2 µL) | ANTIBODY AGAINST NEGATIVE MARKER (2 µL) | TOTAL VOLUME OF ANTIBODIES | VOLUME OF CELL SUSPENSION ADDED | VOLUME OF STAINING BUFFER ADDED | |
| Mixed tube 1 | - Anti-human CD44-V450 | Anti-human CD45-PE | 10 µL | 50 µL | 40 µL | |
| - Anti-human CD90-FITC | ||||||
| - Anti-human CD73-PerCP Cy5.5 | ||||||
| - Anti-human CD105-APC | ||||||
| Mixed tube 2 | - Anti-human CD44-V450 | Anti-human CD45-PE | 10 µL | 50 µL | 40 µL | |
| - Anti-human CD90-FITC | ||||||
| - Anti-human CD73-PerCP Cy5.5 | ||||||
| - Anti-human CD105-APC | ||||||
Table 6: Sample tubes for multicolor immunophenotyping of SHEDs. Abbreviations: SHEDs = stem cells from human exfoliated deciduous teeth; PE = phycoerythrin.
| TUBE TYPE | ANTIBODY AGAINST POSITIVE MARKER (3 µL) | ANTIBODY AGAINST NEGATIVE MARKER (3 µL) | TOTAL VOLUME OF ANTIBODIES | VOLUME OF CELL SUSPENSION ADDED | VOLUME OF STAIN BUFFER ADDED | |
| Mixed tube 1 | - Anti-human CD90-FITC | Anti-human CD45-PE | 9 µL | 50 µL | 41 µL | |
| - Anti-human CD73-PerCP Cy5.5 | ||||||
| Mixed tube 2 | - Anti-human CD90-FITC | Anti-human CD45-PE | 9 µL | 50 µL | 41 µL | |
| - Anti-human CD73-PerCP Cy5.5 | ||||||
Table 7: Single-cell sorting reaction tubes. Abbreviations: FITC = fluorescein isothiocyanate; PE = phycoerythrin; PerCP = peridinin-chlorophyll-protein.
5. Single-cell sorting
- Preparation of cell sorter
- Install a 100 µm nozzle in the sorter.
NOTE: The appropriate nozzle is at least five times the diameter of the particle to be sorted. The sheath fluid to be used for sorting needs to be decided based on the sample type and the sensitivity of the experiment; for this experiment, proprietary sheath fluid has been used. - Perform the daily instrument quality check (QC) and set up the sorter for the experiment. Refer to the instrument manual for a detailed guide to instrument setup.
- Install a 100 µm nozzle in the sorter.
- Setting up the compensation matrix
- Set the compensation matrix using single-stain compensation tubes from step 4.2.
- In the proprietary software, select Experiment from the Toolbar and click on Compensation setup. Open Create compensation controls.
- Check the markers and confirm. A new specimen is added named "Compensation" under which new tubes named the marker controls are added automatically by the software.
- Select the Unstained tube and run it, to record 5,000 events. Drag the gate to the population of cells and apply it to all compensation controls. This is to set the voltages and negative gate for each fluorescent parameter.
- Similarly, load the single-stain compensation tubes separately, and record and save the data. Select the gate demarcating the population of interest and apply to all compensation controls. This is to set the positive gates for each fluorescent parameter.
- Select Experiment from the Toolbar and click on Calculate compensation values | link and save.
NOTE: Once the auto-comp matrix is generated using the software, the voltage parameters of the fluorochromes cannot be changed for any of the channels in the Mixed tubes.
- Data acquisition
- Record 10,000 cells in each tube from steps 4.3. and 4.4 to collect data for analysis of the immunophenotype of the cells.
- Preparation of the collection devices
- Depending on the purpose of the sorted cell populations, choose between 6-well, 24-well, 48-well, or 96-well plates.
- Coat the wells with 200-500 µL of FBS and keep the plates undisturbed for 2 h.
- After 2 h, remove the residual FBS and add 200-500 µL of 10% MSC culture media.
- Cell sorting in single-cell sort mode
- Run Mixed tube 1 (from step 4.5), and record 10,000 events to set the gates on the population of interest to be sorted, using the appropriate gating strategy.
- Load a collection plate and set target cell numbers between 2,500 and 5,000 cells/well and select the single-cell sort purity mask.
- Collect the sorted populations in the collection device and keep them on ice until the end of the sorting experiment.
- Once done, transfer the plates to the 5% CO2 incubator to maintain the cultures at 37 °C.
NOTE: Post acquisition, raw data files were exported as .fcs file format (v.3.0. onwards). The sort reports generated after every experiment recorded the number of events/cells sorted per assigned well and indicated the number of conflicts that were aborted.
Results
The SHEDs were characterized with standard immunofluorescence assays showing the expression of vimentin (red, type III intermediate filaments), actin filaments (Alexa fluor 488 Phalloidin Probes), and nuclei stained with DAPI (Figure 1A). To estimate their proliferative and colony-forming capacities, standard short-term cell growth assays were performed. A 14.3-fold increase in proliferation rate from day 2 to day 8 has been shown in Figure 1B. The clonogenic pr...
Discussion
In the field of tissue engineering and regenerative medicine, among the postnatal sources, oral tissue-derived MSCs have attracted profound interest because of their minimal ethical obligations and notable multilineage differentiation potential21. Dental pulp stem cells (DPSCs) from the impacted third molar and SHEDs have garnered the most attention among dental MSCs for their therapeutic potential in neurodegenerative and traumatic diseases22. The protocol described in thi...
Disclosures
The authors declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgements
We thank the Flow Cell Facility at Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru, India, for the use of the flow cytometry core facility. The cryo-sectioning of the pellet culture of differentiated cells was performed at Neuberg Anand Reference Laboratory, Bengaluru, India. This work was supported by UC's intramural funding from the Manipal Academy of Higher Education (MAHE), India. AG is grateful for the support of the Dr. T. M. A. Pai Scholarship from MAHE.
Materials
| Name | Company | Catalog Number | Comments |
| Alcian Blue Stain | HiMedia | CCK029-1KT | |
| Antibiotic-Antimycotic (100x) | Gibco by ThermoFisher | 15240062 | |
| BD CompBead Plus Anti-Mouse Ig, κ/Negative Control (BSA) Compensation Plus (7.5 µm) Particles Set | BD Biosciences | 560497 | |
| BD FACS Accudrop Beads | BD Biosciences | 345249 | Used to set up the Laser delay when the sort module opens. |
| BD FACS Aria Fusion Flow cytometer | BD Biosciences | --- | |
| BD FACS Diva 9.4 | BD Biosciences | --- | |
| BD FACS Sheath Fluid | BD Biosciences | 342003 | Used as sheath fluid for both analysis and sorting experiments in the BD FACSAria Fusion |
| BD FACSDiva CS&T Research Beads | BD Biosciences | 655050 | Used for Instrument configuration depending on the nozzle size. |
| BD Horizon V450 Mouse Anti-Human CD44 | BD Biosciences | 561292 | |
| BD Horizon V450 Mouse IgG2b, κ Isotype Control | BD Biosciences | 560374 | CD44-V450 isotype |
| BD Pharmingen APC Mouse Anti-Human CD105 | BD Biosciences | 562408 | |
| BD Pharmingen APC Mouse IgG1, κ Isotype Control | BD Biosciences | 555751 | CD105-APC isotype |
| BD Pharmingen DAPI Solution | BD Biosciences | 564907 | DAPI Stock solution of 1 mg/mL |
| BD Pharmingen FITC Mouse Anti-Human CD90 | BD Biosciences | 555595 | |
| BD Pharmingen FITC Mouse IgG1, κ Isotype Control | BD Biosciences | 555748 | CD90-FITC isotype |
| BD Pharmingen PE Mouse Anti-Human CD45 | BD Biosciences | 555483 | |
| BD Pharmingen PE Mouse IgG1, κ Isotype Control | BD Biosciences | 555749 | CD45-PE isotype |
| BD Pharmingen PerCP-Cy 5.5 Mouse Anti-Human CD73 | BD Biosciences | 561260 | |
| BD Pharmingen PerCP-Cy 5.5 Mouse IgG1, κ Isotype Control | BD Biosciences | 550795 | CD73-PerCP-Cy 5.5 isotype |
| BD Pharmingen Purified Mouse Anti-Vimentin | BD Biosciences | 550513 | |
| Bovine serum albumin | Hi-Media | TC548-5G | |
| Crystal violet | Nice chemical pvt ltd | C33809 | |
| Dulbecco's Phosphate Buffered Saline | Sigma-aldrich | D5652-50L | dPBS used for culture work and maintenance. |
| Ethanol | --- | --- | Used for general sterlization. |
| Fetal Bovine Serum | Gibco by ThermoFisher | 10270-106 | |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | ThermoFisher Scientific | A-21422 | |
| KO-DMEM | Gibco by ThermoFisher | 10829018 | Basal medium for undifferentiated hESCs, used in the preparation of culture media |
| L-Glutamine 200mM (100x) | Gibco by ThermoFisher | 25030-081 | |
| Methanol, for Molecular Biology | Hi-Media | MB113 | |
| Oil red O | HiMedia | CCK013-1KT | |
| Paraformaldehyde | loba chemie | 30525-89-4 | |
| Penicillin Streptomycin (100x) | Gibco by ThermoFisher | 15140- 122 | |
| Phalloidin (ActinGreen 488 ReadyProbes reagent) | Invitrogen | R37110 | |
| Silver Nitrate | HiMedia | MB156-25G | |
| Sodium Thiosulphate pentahydrate | Chemport | 10102-17-7 | |
| Sphero Rainbow Fluorescent Particles, 3.0 - 3.4 µm | BD Biosciences | 556291 | |
| Staining buffer | Prepared in MIRM | ---- | It was prepared using 2% FBS in PBS |
| StemPro Adipogenesis Differentiation Basal Media | Gibco by ThermoFisher | A10410-01 | Basal media for Adipogenic media |
| StemPro Adipogenesis Supplement | Gibco by ThermoFisher | A10065-01 | Induction media for Adipogenic media |
| StemPro Chondrogenesis Supplement | Gibco by ThermoFisher | A10064-01 | Induction media for Chondrogenic media |
| StemPro Osteogenesis Supplement | Gibco by ThermoFisher | A10066-01 | Induction media for Osteoogenic media |
| StemPro Osteogenesis/Chondrogenesis Differentiation Basal Media | Gibco by ThermoFisher | A10069-01 | Basal media for both Ostegenic and Chondrogenic media |
| Triton-X-100 | Hi-Media | MB031 | |
| Trypan Blue | Gibco by life technologies | 15250-061 | |
| Trypsin - EDTA Solution 1x | Hi-media | TCL049 | |
| Tween-20 | MERCK | 9005-64-5 |
References
- Kobolak, J., Dinnyes, A., Memic, A., Khademhosseini, A., Mobasheri, A. Mesenchymal stem cells: Identification, phenotypic characterization, biological properties and potential for regenerative medicine through biomaterial micro-engineering of their niche. Methods. 99, 62-68 (2016).
- Wilson, A., Hodgson-Garms, M., Frith, J. E., Genever, P. Multiplicity of mesenchymal stromal cells: finding the right route to therapy. Frontiers in Immunology. 10, 1112 (2019).
- Li, J., et al. Comparison of the biological characteristics of human mesenchymal stem cells derived from exfoliated deciduous teeth, bone marrow, gingival tissue, and umbilical cord. Molecular Medicine Reports. 18 (6), 4969-4977 (2018).
- McLeod, C. M., Mauck, R. L. On the origin and impact of mesenchymal stem cell heterogeneity: new insights and emerging tools for single cell analysis. European Cells & Materials. 34, 217-231 (2017).
- Dominici, M., et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 8 (4), 315-317 (2006).
- Wang, J., Liao, L., Wang, S., Tan, J. Cell therapy with autologous mesenchymal stem cells-how the disease process impacts clinical considerations. Cytotherapy. 15 (8), 893-904 (2013).
- Kern, S., Eichler, H., Stoeve, J., Kluter, H., Bieback, K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 24 (5), 1294-1301 (2006).
- Dunn, C. M., Kameishi, S., Grainger, D. W., Okano, T. Strategies to address mesenchymal stem/stromal cell heterogeneity in immunomodulatory profiles to improve cell-based therapies. Acta Biomaterialia. 133, 114-125 (2021).
- Yang, Y. K., Ogando, C. R., Wang See, C., Chang, T. Y., Barabino, G. A. Changes in phenotype and differentiation potential of human mesenchymal stem cells aging in vitro. Stem Cell Research & Therapy. 9 (1), 131 (2018).
- Costa, L. A., et al. Functional heterogeneity of mesenchymal stem cells from natural niches to culture conditions: implications for further clinical uses. Cellular and Molecular Life Sciences. 78 (2), 447-467 (2021).
- Bonner, W. A., Hulett, H. R., Sweet, R. G., Herzenberg, L. A. Fluorescence activated cell sorting. Review of Scientific Instruments. 43 (3), 404-409 (1972).
- Schulte, R., et al. Index sorting resolves heterogeneous murine hematopoietic stem cell populations. Experimental Hematology. 43 (9), 803-811 (2015).
- Liao, X., Makris, M., Luo, X. M. Fluorescence-activated cell sorting for purification of plasmacytoid dendritic cells from the mouse bone marrow. Journal of Visualized Experiments. (117), (2016).
- Roda, B., et al. A novel stem cell tag-less sorting method. Stem Cell Reviews and Reports. 5 (4), 420-427 (2009).
- Hall, S. R., et al. Identification and isolation of small CD44-negative mesenchymal stem/progenitor cells from human bone marrow using elutriation and polychromatic flow cytometry. Stem Cells Translational Medicine. 2 (8), 567-578 (2013).
- Eggleton, M. J., Sharp, A. A. Platelet counting using the Coulter electronic counter. Journal of Clinical Pathology. 16 (2), 164-167 (1963).
- Porwit-Ksiazek, A., Aman, P., Ksiazek, T., Biberfeld, P. Leu 7+ (HNK-1+) cells. II. Characterization of blood Leu 7+ cells with respect to immunophenotype and cell density. Scandinavian Journal of Immunology. 18 (6), 495-449 (1983).
- Hewitt, Z., et al. Fluorescence-activated single cell sorting of human embryonic stem cells. Cloning and Stem Cells. 8 (3), 225-234 (2006).
- Singh, A. M. An efficient protocol for single-cell cloning human pluripotent stem cells. Frontiers in Cell and Developmental Biology. 7, 11 (2019).
- Wilson, N. K., et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell. 16 (6), 712-724 (2015).
- Zhou, L. L., et al. Oral mesenchymal stem/progenitor cells: the immunomodulatory masters. Stem Cells Inernational. 2020, 1327405 (2020).
- Fawzy El-Sayed, K. M., et al. Adult mesenchymal stem cells explored in the dental field. Advances in Biochemical Engineering/Biotechnology. 130, 89-103 (2013).
- Hardy, W. R., et al. Transcriptional networks in single perivascular cells sorted from human adipose tissue reveal a hierarchy of mesenchymal stem cells. Stem Cells. 35 (5), 1273-1289 (2017).
- . . FACSAria Fusion User's Guide. , (2018).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved