A subscription to JoVE is required to view this content. Sign in or start your free trial.
Improving Student Outcomes with an Adaptable Molecular Cloning Course-Based Undergraduate Research Experience
* These authors contributed equally
In This Article
Summary
An adaptable Gibson Assembly molecular cloning module was employed in a course-based undergraduate research experience (CURE) format for molecular biology laboratory course students. Assessment of student learning outcomes showed improved understanding and confidence in molecular cloning after completion of the CURE, and novel plasmids were cloned for natural product biosynthesis research.
Abstract
The continuous advancement of molecular biology techniques requires that molecular biology curricula are regularly refined to effectively prepare students to enter the workforce with modern competencies. In particular, the emergence of Gibson Assembly, a highly customizable and adaptive molecular cloning technique, has advanced the landscape of molecular cloning in numerous research environments. Thus, we created a Gibson Assembly cloning module for deployment in a molecular biology laboratory course at California Polytechnic State University, San Luis Obispo and evaluated student learning outcomes from the module. Over three iterations of the course, students participated in an experiment-based independent project that involved cloning three unique plasmid libraries to support research projects in natural products biosynthesis. Students were given pre- and post-questionnaires to evaluate their understanding of molecular cloning and their confidence in molecular biology terms and techniques. Students’ responses showed a significant increase in both learning molecular cloning concepts and in self-reported confidence with molecular cloning terms and techniques. This module framework can be generalized to teach Gibson Assembly for various applications, providing instructors with a toolkit for teaching an adaptable and emergent cloning technology while advancing their research projects.
Introduction
Training undergraduates in fundamental molecular biology concepts and laboratory techniques is crucial for their scientific and professional development as these methodologies are common in various research settings, including academia and industry. As such, students in the biology (molecular and cellular biology concentration) and biochemistry majors at California Polytechnic State University, San Luis Obispo (Cal Poly) are required to take an upper-division molecular biology laboratory course to learn and apply these topics (CHEM/BIO 475). A base curriculum for this course has been previously developed wherein students perform topoisomerase-based (TOPO) cloning to assemble an actin-containing plasmid that is prepared from a yeast complementary DNA (cDNA) template1. Students design experiments based on questions that mimic authentic research hypotheses, increasing their familiarity with laboratory practices and inquiry-based learning. Continuous advancement in the field of molecular biology requires that corresponding curricula adapt to prepare students with modern competencies for the workforce. In particular, the use of Gibson Assembly has become more predominant in the scientific community; while the method was originally established to synthesize artificial chromosomes2, over 5,000 publications at the time of this report have referenced Gibson et al.’s original work. Gibson Assembly is unique compared to traditional cloning methodologies: it is highly customizable and can easily ligate multiple linear DNA fragments without the need for restriction sites to produce the junctions. Thus, we saw the opportunity to revamp the CHEM/BIO 475 curriculum to incorporate modern molecular cloning techniques and improve the inquiry-based course model.
It has been established that student research experiences contribute to increased conceptual understanding, skill development, and persistence in science3, yet not all undergraduate students have the opportunity to participate directly in a research laboratory. To address the challenge of limited student capacity in research laboratories, course-based undergraduate research experiences (CUREs) have been developed and employed to increase science accessibility through authentic research in the classroom. While CUREs vary in their implementation, common practices that address the five tenets of scientific research have been established. In a well-designed CURE, students will 1) use scientific practices, 2) collaborate in a research project, 3) attempt to make new discoveries, 4) contribute to work relevant outside of the classroom, and 5) reassess and revise hypotheses and methods in the case of experimental failure4. Similar to traditional student research experiences in a laboratory, CUREs have been shown to strengthen student confidence in science, scientific skills, project ownership, and persistence in science, technology, engineering, and mathematics (STEM)5. While CUREs involving molecular cloning have been reported previously6,7,8,9,10,11,12,13, we are unaware of any that emphasize the adaptability of Gibson Assembly to make a library of authentic research plasmids.
Here, we report an expansion of the current inquiry-based CHEM/BIO 475 curriculum at California Polytechnic State University, San Luis Obispo with two major improvements: hands-on experience using Gibson Assembly and student participation in a CURE, which has provided original plasmid constructs for research projects funded by the National Science Foundation (NSF-1708919 and NSF-2300890). Over three implementations of this curriculum, students have contributed to two distinct research projects focused on natural product biosynthesis of bioactive molecules produced by Actinomycetota. Natural products often contain pharmacophores with antibiotic, antifungal, and/or anticancer activities, giving these small molecules importance in drug discovery efforts and potential for clinical relevance14. This research requires the creation of plasmid libraries to enable investigations of both the function and engineering potential of bacterial biosynthetic enzymes. In this CURE, students designed and performed Gibson Assembly experiments to clone the unique plasmid libraries pertinent to these research projects (Figure 1). In addition, the format and design of the module is distinctive because it is easily adaptable to generate any plasmids of interest for other research projects.
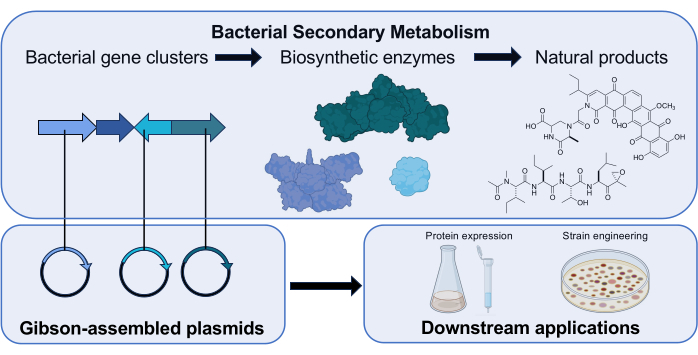
Figure 1: Overview of the role of Gibson Assembly in our research laboratory. Actinomycetota produce small molecule natural products with clinically relevant bioactivities using gene clusters that encode biosynthetic enzymes. In our research, plasmids containing a biosynthetic gene are assembled via Gibson Assembly for downstream investigations of the encoded enzyme’s function. Science icons from Biorender.com. Please click here to view a larger version of this figure.
Laboratory overview
Molecular Biology Laboratory is an upper-division course required for biochemistry majors and biology majors with a molecular and cellular biology concentration. Other students meeting prerequisites are welcome to take the course as an upper-division elective. The course is co-listed between the Chemistry and Biochemistry Department and the Biology Department at Cal Poly. Faculty from both departments take turns teaching the course each term (two quarters Biology, one quarter Chemistry and Biochemistry).
The class meets in the laboratory twice each week for 170 min periods and once a week for a 50 min lecture. Lab sections contain a maximum of 16 students and 2–3 lab sections are offered each quarter. The course runs for 10 weeks, and the final exam is administered during the last meeting in week 10. Lecture time is spent discussing the theory behind many of the experimental techniques conducted in the lab, as well as current topics in molecular biology that are not covered in the lab. The core curriculum of the lab encompasses the process of cloning the actin gene from yeast1, which takes approximately 7 weeks (13–14 lab meetings). Techniques include micropipetting, yeast RNA isolation, amplifying a yeast gene using reverse transcription-polymerase chain reaction (RT-PCR), TOPO cloning, blue-white screening, plasmid isolation and verification of insert by restriction digest and PCR, in silico analysis of clones, and DNA sequence analysis. The curriculum for the last 3 weeks of the course is at the discretion of the instructor but involves students completing an “independent project” with undefined outcomes.
Experiment overview
One focus of our research group is biosynthetic pathways in Actinomycetota. When designing the independent project, we envisioned having students create plasmids through Gibson Assembly for use in our research projects investigating natural product biosynthesis. While the iterations of the course module that are assessed here were particular to plasmids that allow for the manipulation of biosynthetic pathways, the Gibson Assembly workflow is enormously adaptable for other molecular cloning projects (Figure 2). The workflow was split into three different experiments (A, B, and C) that were completed over two lecture periods and six lab periods (3 weeks total) (See Supplemental File 1 and Supplemental File 2). Experiments were preceded by worksheets to support student preparation and assess student understanding (Supplemental File 3, Supplemental File 4, and Supplemental File 5). The workflow is presented in a format that is flexible to an instructor’s needs and interests.
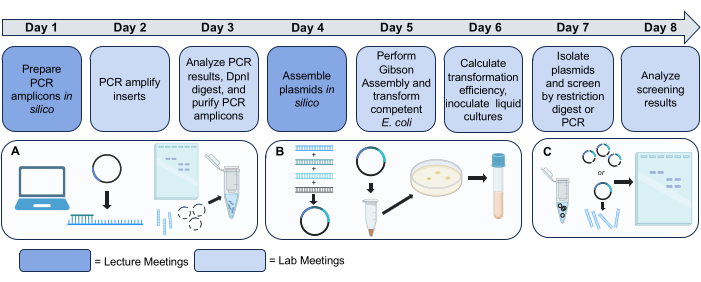
Figure 2: Gibson Assembly module workflow. Day 1 and Day 4 are lecture periods where students complete in silico sequence analysis and experimental design. Days 2–3 and 5–8 are lab meetings where the steps to clone novel plasmids via Gibson Assembly are conducted, followed by isolation and screening. The pictorial flowchart is grouped by the three experiments students perform (A, B, and C). More detailed directions and protocols can be found in the Instructor and Student Manuals provided as Supplemental File 1 and Supplemental File 2, respectively. Science icons from Biorender.com. Please click here to view a larger version of this figure.
The Gibson Assembly independent project module was first piloted in the spring quarter of 2019 in CHEM/BIO 475. In 2020 and 2021, the course was taught online due to the SARS-CoV-2 pandemic. When in-person instruction resumed in the Spring of 2022 and 2023, students within the course were invited to participate in a study assessing learning outcomes of a Gibson Assembly independent project where original, research-relevant plasmids would be cloned. In 2019, students created a library of plasmids consisting of gene cassettes from the genome of Micromonospora echinospora ATCC 15837 that were cloned into pKC1132 (Figure 3). This plasmid library is being utilized in our research lab to inactivate genes of interest in a putative biosynthetic gene cluster for the natural product TLN-0522015. To complement our gene inactivation studies, students in 2022 cloned a small library of genes from the putative TLN-05220 gene cluster into pUC19 (Figure 3); our research group has utilized these plasmids for subcloning genes into expression vectors, including pET28b, for protein overexpression and purification. Students in the 2023 cohort contributed to ongoing work on a biosynthetic engineering project on the epoxomicin synthetase16. In teams of 3–4, students cloned engineered domains of non-ribosomal peptide synthetase17 modules into various protein expression vectors to optimize the overexpression and purification of these enzymes in our research lab (Figure 3). Redundancy was built into the cloning plan for each cohort. For example, the 2019 cohort contained 44 students, and 15 plasmids were assigned to the class, for cloning. Thus, cloning of each plasmid was attempted two or three times.
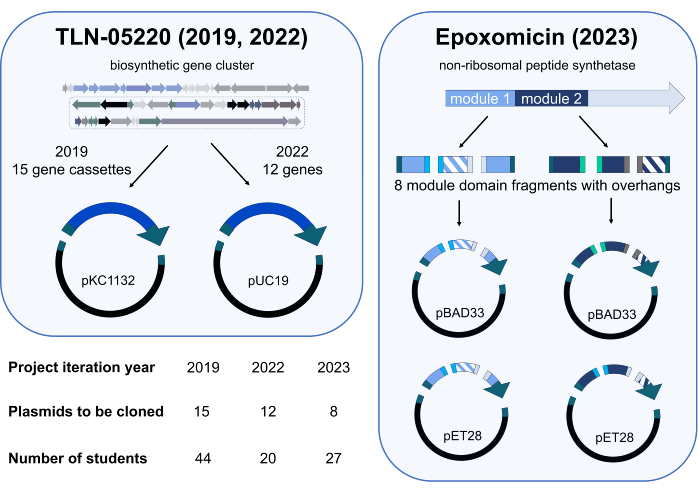
Figure 3: Summary of plasmids cloned and student participants during 2019, 2022, and 2023 independent project iterations. The Gibson Assembly project has been deployed three times. In each offering, student participants cloned a different library of plasmids to be utilized in research projects exploring biosynthetic pathways. Projects in 2019 and 2022 supported our ongoing work on the natural product TLN-0522015 with two fragment (one gene cassette or gene and a vector) Gibson Assembly reactions. The 2023 project involved domain swapping within modules 1 and 2 of a non-ribosomal peptide synthetase (NRPS) enzyme that is involved in epoxomicin biosynthesis16. The hatched fragments represent two different mutants of the swapped domain, and solid colors represent domains that were not swapped. In total, eight different gene fragments (four for module 1 and four for module 2) were generated with overhangs compatible for Gibson Assembly. For each module, two different combinations of three gene fragments were assembled with one of two different vectors (pBAD33 and pET28, four total fragments per assembly), for the potential to generate eight engineered NRPS plasmids. Science icons from Biorender.com. Please click here to view a larger version of this figure.
Assessment overview
At least 1 week prior to the start of the Gibson Assembly independent project, students in the 2022 and 2023 cohorts were invited to participate in a learning outcome investigation, designed similarly to a study performed in a survey of biochemistry lab course at Cal Poly18. Student participants completed a multiple-choice pre-questionnaire in the lab meeting before the start of the independent project and a multiple-choice post-questionnaire during the final lab meeting (i.e., after they had completed the Gibson Assembly module). The pre- and post- questionnaires consisted of 28 identical questions with two additional questions in the post-questionnaire (30 total). Ten content questions were written to assess students’ knowledge of enzymes and mechanisms involved in molecular cloning (e.g., polymerase chain reaction [PCR], Gibson Assembly, transformation, blue-white screening). The following seven questions asked students to self-assess their familiarity with molecular cloning terms (e.g., DNA polymerase, exonuclease, ligase). The next 10 questions allowed students to self-assess their ability to perform molecular cloning techniques (e.g., DNA sequence analysis, restriction digest reactions, agarose gel electrophoresis). Students also reported whether they would be comfortable pursuing a career in molecular biology based on their knowledge of molecular cloning techniques. Two additional questions were included in the post-questionnaire for students to self-assess their commitment to learning in the course and whether the course was a valuable learning experience (Supplemental File 6 and Supplemental File 7). All data from the 2022 and 2023 student pre- and post-questionnaire responses were combined for analysis and are available in Supplemental Table S1.
Protocol
The studies involving human participants were reviewed and approved by human subjects in the Research Institutional Review Board at Cal Poly (2022-113-CP (IRB)). The participants provided their written informed consent to participate in this study.
The following protocol outlines instructor preparation (steps 1.1–1.3), students’ actions for a three-experiment teaching module that includes PCR to obtain linear fragments (steps 2.1–2.7), Gibson assembly, transformation, and selection of clones (steps 3.1–3.5), plasmid isolation and screening (steps 4.1–4.4), and assessment of learning outcomes (5.1–5.2). Instructor preparation describes one representative example of primer design and preparation of a desired plasmid map in silico. All sections of the protocol are adaptable for other desired plasmids. Each student experiment is divided into two 3 h lab meetings.
1. Instructor preparation
- Gibson Assembly primer design using Benchling Assembly Wizard
- Determine the DNA template source for the gene insert(s), such as genomic DNA, plasmid and synthetic DNA.
NOTE: For genes that will be amplified from complex genomic DNA templates, it is advised to first determine unique binding sites for primers within the genomic DNA template that are specific for the gene(s) of interest, using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST)21, as detailed in Supplemental File 1. For simple templates such as plasmids or synthetic DNA, proceed to the next step. - Retrieve DNA sequences of the gene insert(s) and vector (for example, see polyketide hydroxylase gene sequence and pUC19 sequence provided as GenBank files in the Supplemental File 8). Import the desired insert sequence(s) and desired vector sequence to Benchling, each as a new DNA sequence file. Open each imported sequence to be included in the Gibson Assembly reaction.
NOTE: One can import DNA sequence files in several formats, including GenBank and FASTA. One can also import a sequence directly from a database with an accession number or paste in a nucleotide sequence copied from another file or viewer. - At the bottom of the screen, locate the Assembly Wizard tool. Click Assembly Wizard, then select Create New Assembly. From the options provided, select Gibson and click Start to begin the assembly.
- In the vector sequence window, select all bases to be included in the assembly from the vector backbone. Start the selection at the nucleotide position at the 3’ end of the gene insert (e.g., position 657 of pUC19), and select all remaining nucleotides to be included (e.g., through position 656 to include entire pUC19 sequence). Once selected, click the Backbone tab at the bottom of the screen| Set Fragment.
NOTE: Toggle between the Sequence Map and Plasmid views of the vector to select all desired bases of a circular template. Alternatively, if planning to prepare the vector fragment via restriction digest (instead of PCR) for Gibson Assembly, linearize the backbone sequence with a restriction enzyme cut site (e.g., XbaI). To do so, click the cut site, hold Shift, and click the cut site again. Then click Set Fragment to set the backbone. Be aware that vector-specific primers will not be generated by Benchling if the backbone is linearized with a restriction enzyme in the Assembly Wizard. - In the insert sequence window, select all the insert's bases to be included in the assembly. Once selected, click the Insert tab at the bottom of the screen | Set Fragment.
- In case of multiple gene inserts, click the + button on the right side of the Assembly Wizard. In the insert sequence window, select all the insert bases to be included in the assembly. Once selected, click the Insert tab at the bottom of the screen | Set Fragment.
- Once all fragments are set, rename the assembly to the desired plasmid name and click Assemble on the right of the Assembly Wizard. Select the desired folder location for both the Sequence Folder and Primer Folder. Click Create to assemble the recombinant plasmid sequence.
- Open the assembled plasmid and click Assembly History to view a basic plasmid map and an overview of the sequences from which it was derived. Ensure that the nucleotide positions for each fragment match the desired design.
- In the Assembly Parameters tab, view the names of the primers designed for the assembly. Note that there will be two primers designed for each insert (a forward and a reverse), and the primer names will be derived from the titles of the DNA sequence files. Also ensure that primer melting temperatures and suggested annealing temperatures are compatible in this window (see Supplemental File 8, Representative Primers).
- If primer melting temperatures or annealing temperatures are not ideal, manually adjust primer sequences (e.g., removing nucleotides to reduce melting temperature) within the primer sequence files. Alternatively, click on Re-open and adjust the primer settings by clicking the Enzyme/Primer Settings button next to the Assemble button in the Assembly Wizard. Then, repeat the Assembly step and re-examine primer sequences.
- If a library of plasmids is to be cloned with many different genes cloned into the same vector backbone,
- Utilize the same primer pair for the vector in each case. To do so, manually remove all the nucleotides from the vector primers that would install the overhangs that are specific to the insert. These nucleotides would be found at the 5’ end of the primer. After removal, ensure that primer melting temperatures and annealing temperatures are compatible (see Supplemental File 8, Representative Primers).
- Ensure all insert primer pairs have very similar melting temperatures and annealing temperatures.
NOTE: This will reduce the need for annealing temperature optimization of every pair and allows for all student PCR reactions to be cycled at the same time using the same thermocycler program.
- Once primer design is complete for desired plasmids, click Finalize in the Assembly Wizard. Order the primers from a DNA synthesis company and retrieve samples of the DNA templates for insert(s) and vector.
- Test both insert- and vector-specific primer pairs for optimal annealing temperatures using a gradient PCR experiment with the DNA templates. Test the Benchling-recommended annealing temperatures ± 3 °C initially.
- Once annealing temperatures have been determined, create aliquots of primer solutions, DNA templates, and necessary reagents for PCR reactions for students.
- Determine the DNA template source for the gene insert(s), such as genomic DNA, plasmid and synthetic DNA.
- Before the experiments, provide worksheets to the students to support preparation and assess understanding (Supplemental File 4, Supplemental 5, and Supplemental 6).
- Data collection on learning outcomes
NOTE: If interested in collecting learning outcome data from the Gibson Assembly module and publish the results, gain approval for research with human subjects per the institution’s policies.- Invite the students to participate in a learning outcome investigation at least 1 week prior to the start of the Gibson Assembly independent project.
- Have the student participants complete the multiple-choice pre-questionnaire (Supplemental File 6) the lab meeting before the start of the independent project.
2. Student experiment A: PCR to obtain linear fragments
- Obtain primers and template DNA solutions for PCR from the instructor.
- Pipette 25 µL reactions including DNA polymerase MasterMix, DNA template, forward and reverse primers, and nuclease-free water according to instructions in the student manual (see Supplemental File 2).
- Cycle the reaction in a thermocycler according to the student manual in Supplemental File 2. Ensure that the annealing temperature of the reaction is appropriate for the primers, and that the extension time is appropriate for the length of the desired amplicon. Refer to Supplemental File 2 and Supplemental File 3 for detailed instructions.
- Store PCR reactions at 4 °C or −20 °C until the following lab period.
- Analyze 5 µL of each reaction via agarose gel electrophoresis. While the gel is running, add 1 µL of DpnI restriction enzyme to each reaction that utilized plasmid DNA as a template. Incubate this mixture for 1 h at 37 °C.
NOTE: DpnI will degrade any remaining circular plasmid DNA template, making it less likely to obtain false positives in the transformation step. - Image the gel to confirm that PCR was successful and correct amplification was achieved. Purify any successful PCR reactions with a commercially available PCR purification kit. Measure the concentration (ng/µL) of purified PCR product using a microvolume spectrophotometer for use in subsequent Gibson assembly calculations.
- Before the next class meeting, design the reaction recipe for Gibson Assembly (see Supplemental File 2 and Supplemental File 4).
3. Student experiment B: Gibson Assembly, transformation, and selection of clones
- Pipette the Gibson Assembly reaction according to the designed recipe. Incubate the reaction at 50 °C for 15 min. While reactions incubate, prepare for transformation by thawing chemically competent Escherichia coli cells on ice.
- Transform 2 µL of the Gibson Assembly into chemically competent cells via heat shock. Pipette 100 µL of the transformed cells on two selection plates and spread with sterilized beads. Prepare 10-4, 10-5, and 10-6 dilutions of the remaining cells on Luria-Bertani (LB) agar. Plate 100 µL of each serial dilution on LB (no selection) and spread with sterilized beads.
NOTE: This step allows for the calculation of transformation efficiency. - Incubate the plates overnight at 37 °C. Store the plates at 4 °C until the next class period.
- Count colonies on all plates and calculate transformation efficiency as instructed in Supplemental File 2. Re-streak one selective plate with four colonies from either selective plate and incubate this plate at 37 °C overnight.
NOTE: This serves as a backup culture of your positive clones. - Using a marker, select, circle, and label four distinct colonies on the selection plate with the student’s initials and a number (e.g., ABC1). Retrieve a new LB-agar plate containing antibiotic for selection, use a marker to divide the plate into quadrants (i.e., fourths), and then use ~1/2 of each colony to re-streak onto the respectively labeled quadrant and the other 1/2 to inoculate a 5 mL liquid LB culture for each of the four colonies selected. Be sure to label the tubes with the respective colony identity (e.g., ABC1) and add the correct concentration of antibiotic for selection. Incubate liquid cultures in a shaking incubator at 37 °C overnight and the agar plate in a static incubator at 37 °C; store liquid cultures and plates at 4 °C until the following class period.
4. Student experiment C: Plasmid isolation and screening
- Isolate plasmid DNA from the liquid cultures from Experiment B using a miniprep kit. Measure the concentration of isolated plasmid (in ng/µL).
- Design a restriction digest or PCR screen to analyze the isolated plasmids (see detailed instructions in Supplemental File 2 and report expectations in Supplemental File 5).
- Pipette the restriction digest reactions or reactions according to designed recipes and incubate at temperatures and durations designed earlier.
- Analyze results via agarose gel electrophoresis.
5. Assessment
- Have the student participants complete the multiple-choice post-questionnaire (Supplemental File 7) the lab meeting after the Gibson Assembly module is complete.
- Combine all data from the pre- and post-questionnaire responses for analysis.
- Examine the statistical significance of the increase in the average score of each individual content question between the questionnaires by using two-tailed paired t-tests and Cohen’s d effect size.
- Evaluate the student confidence increase for each individual term and technique to determine the extent of change and its statistical significance.
- Determine the impact of student background and academic factors on student learning by using normalized learning gain (NLG) and unpaired t-tests.
- Assess the students’ attitudes towards the project experience and molecular biology careers by evaluating the average scores.
Results
Student success in cloning
In each iteration of the Gibson Assembly module (2019, 2022, and 2023), students were asked to prepare a report summarizing their findings. In 2019, 36 of 44 students (81.8%) reported that they successfully cloned their plasmids based on the results of the screen they designed for Experiment C. A total of 14 out of 20 students (70.0%) reported success in cloning their assigned constructs in 2022, while the team-based project in 2023 had 12 of 27 students (44.4% or 4 out o...
Discussion
Here, we propose an adjustable undergraduate classroom laboratory project that teaches students molecular cloning through Gibson Assembly in a course-based research setting. In total, 28 novel plasmids were cloned by undergraduate students in a classroom setting. Individual student-reported success ranged from 44.4% to 81.8% over three cohorts, and overall cloning success was 80% (28 of 35 total assigned plasmids were successfully cloned). Lower cloning success in 2023 was likely due to two compounding factors. First, tu...
Disclosures
The authors declare that they have no competing financial interests or other conflicts of interest.
Acknowledgements
Authors gratefully acknowledge Andrea Laubscher for technical support, and Michael Black, Sandi Clement, and Javin Oza for helpful discussions on teaching lab implementation and assessment of learning outcomes. Authors appreciatively acknowledge all students who participated in the learning outcome study in the 2019, 2022, and 2023 cohorts, as well as research students Nathan Kuhn and Aayushi Adettiwar who assisted in reagent preparation for teaching lab implementation. Authors also acknowledge funding support from the William and Linda Frost Fund, Center for Applications in Biotechnology’s Chevron Biotechnology Applied Research Endowment Grant, and the National Science Foundation (NSF-1708919 and NSF-2300890).
Materials
| Name | Company | Catalog Number | Comments |
| Deoxyribonucleotide triphosphate (dNTPs, 10 mM) | Fisher Scientific | FERR0191 | Homemade' MasterMix component |
| Dithiothreitol (DTT) | Fisher Scientific | FERR0861 | Homemade' MasterMix component |
| DpnI | New England Biolabs | R0176S | 1000 units |
| Fisherbrand Isotemp Microbiological Incubator | Fisher Scientific | 15-103-0513 | |
| FisherBrand Isotemp Water Bath | Fisher Scientific | S28124 | |
| GelRed Nucleic Acid Gel Stain | Biotium | NC9594719 | 10,000X |
| GeneJET Gel Extraction and DNA Cleanup Micro Kit | Thermo Scientific | FERK0831 | 100 Preps |
| GeneRuler 1 kb DNA ladder | Fisher Scientific | FERSM0314 | 100 applications |
| LB Broth, Miller | Fisher BioReagents | BP9723-500 | 500 g |
| Magnesium chloride hexahydrate | Fisher Scientific | BP214-500 | Homemade' MasterMix component |
| Mastercycler nexus X2 Gradient Thermocycler | Eppendorf | 6337000027 | |
| Microfuge 16 Centrifuge | Beckman Coulter | A46474 | |
| Micromonospora echinospora bacteria | American Type Culture Collection | ATCC 15837 | |
| Microwave Oven | General Electric | 2440640 | |
| Molecular Biology Grade Agarose | Fisher BioReagents | BP160-100 | 100 g |
| Nanodrop One Microvolume Spectrophotometer | Thermo Scientific | 13-400-518 | |
| NEB 5-alpha Competent E. coli | New England Biolabs | C2987H | 20 x 0.05 mL |
| NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | E2621S | 10 reactions |
| New Brunswick Innova 40 Benchtop Orbital Shaker | New Brunswick | M1299-0090 | |
| Nuclease Free Water | Fisher BioReagents | BP248450 | 50 mL |
| PEG-8000 | Fisher Scientific | BP233-100 | Homemade' MasterMix component |
| Phusion DNA Polymerase | New England Biolabs | M0530 | Homemade' MasterMix component |
| Portable Balance | Ohaus | SKX123 | |
| pUC19 vector | New England Biolabs | N3041S | |
| Q5 High-Fidelity 2x Master Mix | New England Biolabs | M0492S | 100 reactions |
| T5 Exonuclease | Epicentre | T5E4111K | Homemade' MasterMix component |
| Taq DNA Ligase | New England Biolabs | M0208 | Homemade' MasterMix component |
| Tris-HCl | Fisher Scientific | AAA1137918 | Homemade' MasterMix component |
| TriTrack DNA Gel Loading Dye (6x) | Thermo Scientific | FERR1161 | 5 x 1 mL |
| Zyppy Plasmid Miniprep Kit | Zymo Research | D4019 | 100 Preps |
| β-Nicotinamide adenine dinucleotide (NAD+) | New England Biolabs | B9007S | Homemade' MasterMix component |
References
- Black, M. W., Tuan, A., Jonasson, E. Cloning yeast actin cDNA leads to an investigative approach for the molecular biology laboratory. Biochem Mol Biol Educ. 36 (3), 217–224 (2008).
- Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., Smith, H. O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6 (5), 343–345 (2009).
- Bell, J. K. et al. CUREs in biochemistry—where we are and where we should go. Biochem Mol Biol Educ. 45 (1), 7–12 (2017).
- Auchincloss, L. C. et al. Assessment of course-based undergraduate research experiences: A meeting report. CBE Life Sci Educ. 13 (1), 29–40 (2014).
- Buchanan, A. J., Fisher, G. R. Current status and implementation of science practices in Course-based Undergraduate Research Experiences (CUREs): A systematic literature review. CBE Life Sci Educ. 21 (4), ar83 (2022).
- Verity, N., Ulm, B., Pham, K., Evangelista, B., Borgon, R. Demonstrating core molecular biology principles using GST-GFP in a semester-long laboratory course. Biochem Mol Biol Educ. 50 (1), 55–64 (2022).
- Li, G. et al. CUR(E)ating a new approach to study fungal effectors and enhance undergraduate education through authentic research. Biochem Mol Biol Educ. 52 (1), 6–14 (2024).
- Roecklein-Canfield, J. A., Lopilato, J. Tagging and purifying proteins to teach molecular biology and advanced biochemistry. Biochem Mol Biol Educ. 32 (6), 373–377 (2004).
- Li, C. et al. Directed evolution of glyphosate oxidase and a chemiluminescence system for glyphosate detection: A comprehensive practical laboratory experiment on biotechnology. Biochem Mol Biol Educ. 51 (3), 302–311 (2023).
- Wang, J. T. H., Schembri, M. A., Ramakrishna, M., Sagulenko, E., Fuerst, J. A. Immersing undergraduate students in the research experience: A practical laboratory module on molecular cloning of microbial genes. Biochem Mol Biol Educ. 40 (1), 37–45 (2012).
- Dean, D. M., Wilder, J. A. The “Frankenplasmid” lab: An investigative exercise for teaching recombinant DNA methods. Biochem Mol Biol Educ. 39 (5), 376–383 (2011).
- Bornhorst, J. A., Deibel, M. A., Mulnix, A. B. Gene amplification by PCR and subcloning into a GFP-fusion plasmid expression vector as a molecular biology laboratory course. Biochem Mol Biol Educ. 32 (3), 173–182 (2004).
- Roberts, L. A., Shell, S. S. A research program-linked, course-based undergraduate research experience that allows undergraduates to participate in current research on mycobacterial gene regulation. Front Microbiol. 13, 1025250 (2023).
- Jose, P. A., Maharshi, A., Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol Res. 246, 126708 (2021).
- Banskota, A. H. et al. TLN-05220, TLN-05223, new Echinosporamicin-type antibiotics, and proposed revision of the structure of bravomicins. J Antibiot. 62 (10), 565–570 (2009).
- Schorn, M. et al. Genetic basis for the biosynthesis of the pharmaceutically important class of epoxyketone proteasome inhibitors. ACS Chem Biol. 9 (1), 301–309 (2014).
- Smith, H. G., Beech, M. J., Lewandowski, J. R., Challis, G. L., Jenner, M. Docking domain-mediated subunit interactions in natural product megasynth(et)ases. J Ind Microbiol Biotechnol. 48 (3–4), kuab018 (2021).
- Williams, L. C. et al. The genetic code kit: An open-source cell-free platform for biochemical and biotechnology education. Front Bioeng Biotechnol. 8, 941 (2020).
- Bloodhart, B., Balgopal, M. M., Casper, A. M. A., Sample McMeeking, L. B., Fischer, E. V. Outperforming yet undervalued: Undergraduate women in STEM. PLoS One. 15 (6), e0234685 (2020).
- Farrar, V. S., Aguayo, B.-Y. C., Caporale, N. Gendered performance gaps in an upper-division biology course: Academic, demographic, environmental, and affective factors. CBE Life Sci Educ. 22 (4), ar52 (2023).
- Camacho, C. et al. BLAST+: Architecture and applications. BMC Bioinf. 10 (1), 421 (2009).
- New England BioLabs Inc. Optimization Tips for NEBuilder® HiFi DNA Assembly and NEB® Gibson Assembly. https://www.neb.com/en-us/tools-and-resources/usage-guidelines/optimization-tips-for-nebuilder-hifi-dna-assembly-and-neb-gibson-assembly (2024).
- New England BioLabs Inc. Instruction Manual: Gibson Assembly® Master Mix / Gibson Assembly® Cloning Kit. https://www.neb.com/-/media/nebus/files/manuals/manuale2611.pdf?rev=9db62577a41b4cfda071e21864a6763e (2021).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved