A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
Isolation and Characterization of Exosomes Derived from Mouse Spleen Tissues
* These authors contributed equally
In This Article
Summary
Here, we present a protocol to isolate mouse spleen-derived exosomes using a combination of digestion with collagenase type I and ultracentrifugation.
Abstract
Exosomes (Exo) are lipid-bilayer structures secreted by various cells, including those of animals, plants, and prokaryotes. Previous studies have revealed that Exo derived from humoral or cell-supernatant are promising targets for novel diagnostic or prognostic biomarkers, underscoring their significant role in disease pathogenesis. Tissue-derived Exo (Ti-Exo) have attracted increasing attention due to its ability to accurately reflect tissue specificity and the microenvironment. Ti-Exo, present in interstitial space, play crucial roles in intercellular communication and cross-organ signaling. Despite their recognized value in elucidating disease mechanisms, isolating Ti-Exo remains challenging due to the complexity of tissue matrices and variability in extraction methods. In this study, we developed a practical protocol for isolating exosomes from mice spleen tissue, providing a reproducible technique for subsequent identification analysis and functional studies. We used Type I collagenase digestion combined with differential ultracentrifugation to isolate spleen-derived Exo. The characteristics of isolated Exo were determined through electron microscopy, the nano-flow cytometer, and the western blot. The isolated spleen-derived Exo displayed the typical morphology of lipid bilayer vesicles, with particle sizes ranging from 30 nm to 150 nm. In addition, the expression profile of exosome markers confirmed the presence and purity of exosomes. Taken together, we successfully established a practical protocol for isolating spleen-derived Exo in mice.
Introduction
Exosomes (Exo) are a subgroup of extracellular vesicles (EVs), ranging from 30 to 150 nm in size, encapsulating a diverse array of biomolecules, including proteins, nucleic acids, and lipids1. These biomolecules are derived from the parental cells and are released into the extracellular space. Exo facilitate biomolecular information exchange between cells and their surrounding microenvironment, playing roles in both prokaryotic and eukaryotic systems2. The characteristics of exosomes, such as content, size, membrane components, and cellular origin, are highly variable and influenced by the originating cell type, cellular state, and environmental conditions.
Exosomes are commonly classified into three categories based on their source: cell culture-derived, body fluid-derived, and tissue-derived (Ti-Exo). While cell culture-derived exosomes have been extensively studied due to their accessibility and consistent yield, their use is limited by potential alterations in cell characteristics after prolonged culture, which may misrepresent their biological functions3,4,5. Moreover, most cell cultures are maintained in two-dimensional environments that do not mimic the complex in vivo intercellular interactions, potentially impacting data interpretation6. Conversely, exosomes isolated from biological fluids offer a minimally invasive option to monitor disease progression in real time7. However, these samples often contain a mixture of exosomes from various origins, complicating the accurate identification of their primary source8. Given these challenges, there is an increasing interest in Ti-Exo, which reside in the extracellular interstitium and are key mediators of intercellular signaling.
The spleen plays a crucial role in immune function and maintaining internal homeostasis. Despite existing protocols for isolating Ti-Exo from organs such as the brain, liver, and tumors, practical methods for spleen-derived exosomes are limitedly reported9,10. This study aimed to establish a practical protocol for isolating exosomes from spleen tissue in mice modified from a previous report11. We detail a method involving collagenase digestion followed by ultracentrifugation, which minimizes the disruption of the cell membrane and ensures high purity and yield of spleen-derived Exo. The characteristics of isolated Exo using this established protocol were validated through electron microscopy (TEM), the nano-flow cytometer (Nano-FCM), and western blot, confirming the protocol's efficacy for further experimental research.
Protocol
The samples were obtained from mice, with ethical approval granted by the Ethics Committee of Guangdong Medical University. Detailed descriptions of materials, equipment, and software used in this protocol are provided in the Table of Materials. The details of the preparation prior to extraction are illustrated in Figure 1, while the process of Spleen-Exo extraction and enrichment is described in Figure 2.
1. Preparation
- Pre-cool high-speed and ultra-high-speed centrifuges to 4 °C and set the temperature of a desktop shaker to 37 °C.
- Prepare cell culture dishes (
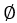 100 mm), 50 mL sterile centrifuge tubes, ice, and sterile cell strainer (
100 mm), 50 mL sterile centrifuge tubes, ice, and sterile cell strainer (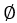 70 µm) as shown in Figure 1A, B.
70 µm) as shown in Figure 1A, B. - Sterilize scissors and forceps using 75% alcohol spray, followed by high-temperature sterilization. These tools are shown in Figure 1C.
- Place the spleen tissue in a sterile
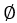 100 mm Petri dish on ice, and wash off the surface blood with 1x iced sterile phosphate-buffered saline (PBS). Dry the spleen tissues with sterile gauze and weigh them after drying.
100 mm Petri dish on ice, and wash off the surface blood with 1x iced sterile phosphate-buffered saline (PBS). Dry the spleen tissues with sterile gauze and weigh them after drying.
NOTE: If the tissue is frozen, thaw it in a dish for 15 min on ice before proceeding. - Moisturize the cell strainer (
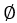 70 µm) by dropping 1 mL of sterile PBS into the strainer using a sterile transfer pipet.
70 µm) by dropping 1 mL of sterile PBS into the strainer using a sterile transfer pipet. - Prepare the digestive buffer (0.1% type I collagenase) using 1x PBS with a vortex mixer.
NOTE: Use 10 mL of digestive buffer for every 100 mg of spleen tissue. The collagenase concentration was chosen in this study based on the product instructions. For other tissue types, researchers may need to adjust these parameters based on tissue density and composition. Preliminary experiments with varying concentrations and digestion times are recommended to determine the optimal conditions for each tissue type.
2. Tissue digestion
- Chop the tissue into uniform small pieces with scissors in a culture dish on ice (Figure 3A) and transfer to a 50 mL centrifuge tube with a sterile transfer pipet.
- Add digestion buffer to the tube according to the spleen tissue weight as indicated above (Figure 3B), then shake the tube with an angle of 45° on a horizontal shaker at 37 °C. Monitor the digestion progress and stop the digestion when tissue chunks have dispersed, and most have lost their shape. Digestion time should vary between tissues. Under the experimental conditions in this study, the digestion time is approximately 30 min.
- Transfer the digested mixture to a cell strainer (Ф70 µm) at room temperature (RT) to remove fibrous and larger tissue debris.
NOTE: Complete filtration immediately after digestion. - Collect the filtrate in a new 50 mL centrifuge tube.
3. Exosome isolation by differential centrifugation
- Centrifuge filtrates at 500 x g for 10 min at 4 °C. A pellet containing residual cells, cell debris, and nuclei can be seen after centrifugation, as shown in Figure 3C.
- Transfer the supernatant to a new tube and centrifuge at 3000 x g for 20 min at 4 °C. A pellet containing apoptotic bodies and microsomes can be seen after centrifugation, as shown in Figure 3D.
- Transfer the supernatant to a new tube and centrifuge at 10,000 x g for 30 min at 4 °C. Pellet containing microvesicles and plasma membrane can be seen after centrifugation, as shown in Figure 3E.
- Ultracentrifuge the resulting supernatant at 120,000 x g for 2 h at 4 °C. A pellet can be seen at the bottom of the tube after centrifugation, as shown in Figure 3F.
- Discard the supernatant, wash the pellet with PBS, and ultracentrifuge again at 120,000 x g for 2 h at 4 °C to obtain exosomes. A pellet can be seen at the bottom of the tube after centrifugation, as shown in Figure 3G.
- Dissolve the isolated exosomes with sterile PBS (200 µL per spleen) and store the isolated exosomes in a new 2 mL centrifuge tube at -80 °C.
4. Exosome characterization by transmission electron microscopy (TEM)
NOTE: TEM was used to determine whether the extracted Exo displayed vesicle characteristics. The Exo samples identified by electron microscopy must be fresh samples or stored briefly at 4 °C for a short time.
- Fix exosomes (2-6 µg/µL) with 2% electron microscopy (EM)-grade paraformaldehyde at RT for 2 h.
- Place 10 µL of exosome solution on a copper mesh, incubate at RT for 10 min, rinse with sterile distilled water, and blot excess liquid with absorbent paper.
- Stain the copper mesh with 2% uranyl acetate for 1 min, blot excess fluid with filter paper, and dry under an incandescent lamp for 2 min.
- Observe the prepared sample under a transmission electron microscope at 80 kV.
5. Size distribution and particle concentration measurement of exosomes
- Load the standard polystyrene nanoparticles (250 nm) to the nano-flow cytometer.
- Measure the protein concentration of Exo samples using a BCA protein assay kit. Dilute Exo sample with PBS according to BCA Protein Assay results (dilute the exosomes to 1-10 ng/µL), then load the Exo samples to the nano-flow to measure the side scatter intensity (SSI).
- Calculate Exo concentration according to the ratio of SSI to particle concentration in the standard polystyrene nanoparticles.
NOTE: For size measurement, a standard curve is generated by loading standard silica nanoparticles with gradient sizes (68 nm, 91 nm, 113 nm, 155 nm) to the nano-flow cytometer. The loading of the Exo sample follows this. The size distribution of Exo was calculated according to the standard cure.
6. Lysis and immunoblot confirmation of exosome proteins
- Add 100 µL of radio immunoprecipitation assay (RIPA) lysis buffer to the exosome pellet for protein analysis.
- Vortex vigorously, cover with parafilm, rock for 20 min at RT, then vortex again.
- Centrifuge briefly at 1000 x g for 30-60 s to collect the sample, transfer to a new 1.5 mL microcentrifuge tube, and store at -20 °C to -80 °C until analysis.
- For immunoblot, mix samples with 5x loading buffer to achieve a 1x concentration and prepare for gel electrophoresis.
- If whole-tissue homogenate controls are desired, use a strong lysis buffer with a protease inhibitor to lyse the tissue, as detailed above. Then, centrifuge the samples at 10,000 x g for 10 min and discard the remaining extracellular matrix, whole cells, or intact cellular debris.
- Boil the sample buffer containing exosomes or tissue homogenate at 95 °C for 5-10 min. Load equal protein per lane into a 10% SDS-PAGE gel. Load equal protein of tissue homogenate as control.
- Proceed with gel electrophoresis under a voltage of 80 V for approximately 2 h using the conventional electrophoresis buffer. Perform western blot analysis to confirm Exo protein expression in the purified lysates and compare relative Exo abundance in fractions.
Results
Isolation and purification of spleen-derived exosomes
To isolate exosomes from mouse spleen tissue, we employed a combination of collagenase digestion and differential ultracentrifugation (Figure 2). The initial steps involved pre-treating the spleen tissue to remove surface blood, followed by digestion with Type I collagenase. This enzymatic treatment facilitated the breakdown of extracellular matrix components, thus liberating the exosomes while preserving their inte...
Discussion
Recent research has reported that spleen-derived exosomes (Spleen-Exo) play a critical role in the treatment of gastric cancer12. To further elucidate the functions of Spleen-Exo, it is necessary to establish a reproducible and optimal method for their extraction from spleen tissue. While protocols exist for isolating exosomes from brain and liver cancer13,14, methods for other tissues remain underdeveloped and require further validation. ...
Disclosures
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82370281, 81870222), the Natural Science Foundation of Guangdong Province (2022A1515012103), and the Science, the Technology Development Special Fund Competitive Allocation Project of Zhanjiang City (2021A05086), and the Startup Foundation from the Second Affiliated Hospital of Guangdong Medical University (23H03).
Materials
| Name | Company | Catalog Number | Comments |
| 70 µm Cell Strainer | Biologix Group Co., Ltd.USA | 15-1070 | |
| Anti CD9 antibody | Cell Signaling Technology, Inc (CST), USA | 983275 | |
| Anti GM130 antibody | Proteintech Group, Inc.China | 66662-1-lg | |
| Anti TSG101 antibody | Abcam Plc, UK | 125011 | |
| Cell Culture Dish | Wuxi NEST Biotechnology Co.,Ltd, China | 704001 | |
| Centrifuge tube | Wuxi NEST Biotechnology Co.,Ltd, China | 788211 | |
| Collagenase Type I | Sigma-Aldrich Corp., USA | C2674 | |
| Desktop Thermostatic Shaker | Shanghai bluepard instruments Co., Ltd., China | THZ-100 | |
| Electrophoresis buffer | Wuhan Servicebio Technology Co., Ltd., China | G2081-1L | |
| Enhanced BCA Protein Assay Kit | Shanghai Beyotime Biotechnology Co., Ltd., China | P0010S | |
| Flow NanoAnalyzer | NanoFCM Inc., China | N30E | |
| Fluorescence/Chemiluminescence imaging system | Guangzhou Biolight Biotechnology Co., Ltd., China | GelView 6000Plus | |
| HRP Goat anti-Mouse IgG | Proteintech Group, Inc.China | 15014 | |
| HRP Goat anti-Rabbit IgG | Proteintech Group, Inc.China | 15015 | |
| microplate reader | Molecular Devices, USA | CMax Plus | |
| Microscissors | Shanghai Medical Instruments (group) Co., Ltd.,China | WA1010 | |
| Microscopic tweezers | Shanghai Medical Instruments (group) Co., Ltd.,China | WA3010 | |
| Multifuge X1R Pro centrifuge | Thermo Fisher Scientific, USA | 75009750 | |
| Ophthalmic scissors | Shanghai Medical Instruments (group) Co., Ltd.,China | Y00030 | |
| Ophthalmic tweezers | Shanghai Medical Instruments (Group) Co., Ltd., China | JD1060 | |
| Optima XPN-100 Ultrafiltration centrifuge | Beckman Coulter, USA | A94469 | |
| phosphate buffered saline (PBS) | Beijing Solarbio Science & Technology Co.,Ltd., China | P1003 | |
| RIPA lysis buffer (strong, without inhibitors) | Shanghai Beyotime Biotechnology Co., Ltd., China | P0013K | |
| Ruler | Deli Manufacturing Company, China | ||
| SDS-PAGE Sample Loading Buffer, 5x | Shanghai Beyotime Biotechnology Co., Ltd., China | P0015 | |
| Transfer Pipet | Biologix Group Co., Ltd.USA | 30-0238A1 | |
| Transmission Electron Microscope | Hitachi, Japan | H-7650 | |
| Ultracentrifugation tube | Beckman Coulter, USA | 355618 | |
| Western Transfer Buffer | Wuhan Servicebio Technology Co., Ltd., China | G2028-1L |
References
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr Biol. 28 (8), R435-R444 (2018).
- Costa-Silva, B., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 17 (6), 816-826 (2015).
- Crescitelli, R., Lasser, C., Lotvall, J. Isolation and characterization of extracellular vesicle subpopulations from tissues. Nat Protoc. 16 (3), 1548-1580 (2021).
- de Jong, B., Barros, E. R., Hoenderop, J. G. J., Rigalli, J. P. Recent advances in extracellular vesicles as drug delivery systems and their potential in precision medicine. Pharmaceutics. 12 (11), 1006 (2020).
- Crescitelli, R., et al. Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. J Extracell Vesicles. 9 (1), 1722433 (2020).
- Jensen, C., Teng, Y. Is it time to start transitioning from 2D to 3D cell culture. Front Mol Biosci. 7, 33 (2020).
- Leung, L. L., Riaz, M. K., Qu, X., Chan, J., Meehan, K. Profiling of extracellular vesicles in oral cancer, from transcriptomics to proteomics. Semin Cancer Biol. 74, 3-23 (2021).
- Shah, R., Patel, T., Freedman, J. E. Circulating extracellular vesicles in human disease. N Engl J Med. 379 (10), 958-966 (2018).
- Wang, X., et al. An enrichment method for small extracellular vesicles derived from liver cancer tissue. J Vis Exp. (192), e64499 (2023).
- Vella, L. J., et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 6 (1), 1348885 (2017).
- Hoshino, A., et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 182 (4), 1044-1061.e18 (2020).
- Chen, Y., et al. Yiwei decoction promotes apoptosis of gastric cancer cells through spleen-derived exosomes. Front Pharmacol. 14, 1144955 (2023).
- Useckaite, Z., et al. Proteomic profiling of paired human liver homogenate and tissue-derived extracellular vesicles. Proteomics. 24 (11), e2300025 (2023).
- Hurwitz, S. N., et al. An optimized method for enrichment of whole brain-derived extracellular vesicles reveals insight into neurodegenerative processes in a mouse model of Alzheimer's disease. J Neurosci Methods. 307, 210-220 (2018).
- Lai, J. J., et al. Exosome processing and characterization approaches for research and technology development. Adv Sci (Weinh). 9 (15), e2103222 (2022).
- Zhang, Y., et al. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomedicine. 15, 6917-6934 (2020).
- Roh, Y. J., et al. Adipose tissue-derived exosomes alleviate particulate matter-induced inflammatory response and skin barrier damage in atopic dermatitis-like triple-cell model. PLoS One. 19 (1), e0292050 (2024).
- Gallart-Palau, X., Serra, A., Sze, S. K. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol Neurodegener. 11 (1), 41 (2016).
- Kurian, T. K., Banik, S., Gopal, D., Chakrabarti, S., Mazumder, N. Elucidating methods for isolation and quantification of exosomes: A review. Mol Biotechnol. 63 (4), 249-266 (2021).
- Menu, E., Vanderkerken, K. Exosomes in multiple myeloma: from bench to bedside. Blood. 140 (23), 2429-2442 (2022).
- Shen, S., et al. Effects of lysate/tissue storage at -80 degrees C on subsequently extracted EVs of epithelial ovarian cancer tissue origins. iScience. 26 (4), 106521 (2023).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved