A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
An Open-Source Framework for Mass Calculation of Antibody-Based Therapeutic Molecules
In This Article
Summary
This article describes the use of a software application, mAbScale, for the calculation of masses for monoclonal antibody-based protein therapeutics.
Abstract
Biotherapeutic masses are a means of verifying identity and structural integrity. Mass spectrometry (MS) of intact proteins or protein subunits provides an easy analytical tool for different stages of biopharmaceutical development. The protein's identity is confirmed when the experimental mass from MS is within a pre-defined mass error range of the theoretical mass. While several computational tools exist for the calculation of protein and peptide molecular weights, they either were not designed for direct application to biotherapeutic entities, have access limitations due to paid licenses, or require uploading protein sequences to host servers.
We have developed a modular mass calculation routine that enables the easy determination of the average or monoisotopic masses and elemental compositions of therapeutic glycoproteins, including monoclonal antibodies (mAb), bispecific antibodies (bsAb), and antibody-drug conjugates (ADC). The modular nature of this Python-based calculation framework will allow the extension of this platform to other modalities such as vaccines, fusion proteins, and oligonucleotides in the future, and this framework could also be useful for the interrogation of top-down mass spectrometry data. By creating an open-source standalone desktop application with a graphical user interface (GUI), we hope to overcome the restrictions around use in environments where proprietary information cannot be uploaded to web-based tools. This article describes the algorithms and application of this tool, mAbScale, to different antibody-based therapeutic modalities.
Introduction
Over the past two decades, biotherapeutics have evolved to become a mainstay of the modern pharmaceutical industry. The SARS-CoV2 pandemic and other life-threatening conditions have further increased the need for the faster and broader development of biopharmaceutical molecules1,2,3.
The biotherapeutic molecular weight is critical for the identification of the molecule, in combination with other analytical assays. The intact and reduced subunit masses are used throughout the discovery and development lifecycles as part of control strategies aimed at maintaining the quality, as described in the QTPP (Quality Target Product Profile)4.
Analytical development in the biopharmaceutical industry relies heavily on mass measurements for intact mass analysis and deep characterization using peptide mapping or multi-attribute method (MAM) monitoring. At the center of these techniques utilizing modern mass spectrometry (MS) platforms is the ability to provide high-resolution accurate mass (HR/AM) measurements. Most HR/AM instruments yield mass accuracies in the range of 0.5-5 ppm, which scale with the mass range. The ability to measure masses accurately for intact large molecules enables the quick and confident identification of large-molecule therapeutics. As isotopic resolution cannot be attained using typical experimental conditions for large molecules (>10 kDa), average masses must be calculated for comparison and identification5,6.
A typical intact or subunit protein mass spectrum represents the overall proteoform profile, which contains composite information on the various molecular forms resulting from post-translational modifications (PTM) and any primary structure differences, such as clips or sequence variants. The relatively easy and high-throughput nature of these measurements make them attractive for characterization and as in-process monitoring controls7,8. Data analysis for these experiments usually requires the user to define the search space for molecular forms (range of PTMs or other molecular forms). For glycosylated proteins, this search space is largely driven by glycoform heterogeneity. Combinations of multiple PTMs, disulfide bond configurations, and other variations along the primary structure make calculating all the possible molecular forms a tedious task. Therefore, the manual calculation of the possible molecular forms is a time and resource-consuming process with a high potential for human error.
Here, we present a mass calculation tool that was developed considering the most important features of biotherapeutic molecules, such as mAbs, bsAbs, ADCs, etc. The tool allows the easy incorporation of search-space variables for the consistent calculation of masses and elemental compositions. The modular nature of this tool will enable it to be further developed and applied to mass calculation and mass matching for other modalities.
The GUI module allows the user to specify the input for the mass calculation, as shown in Figure 1; specifically, the user enters single-letter amino acid sequences for light and heavy antibody chains. Common modifications for heavy-chain N-terminal cyclization and C-terminal lysine clipping are included as check boxes. Further, the chemical formula/elemental composition can be added/subtracted from these protein chains through the respective Chem Mod text box. This allows the user the flexibility to add an elemental composition that includes multiple post-translational modifications or a small-molecule payload in the case of an ADC. As most therapeutic mAbs are engineered to remove the glycosylation sites in the light chain, glycosylation in the light chain is left optional and can be specified using a check box on the GUI.
A typical variation on intact mass analysis for antibodies is a reduced subunit mass analysis, where the light chain is detached from the heavy chain by reducing the interchain disulfide bonds. Depending on the strength of the reducing agent used, the intrachain disulfide bonds may or may not be cleaved. The users have the flexibility of entering the total number of disulfide bonds depending on the IgG subtype or in case of a cysteine-conjugated ADC9.
The application calculates masses in a bottom-up manner, in which the elemental compositions are first calculated for the individual heavy chains and light chains. Next, heavy chain (HC) N-terminal cyclization Lys-clipping is accounted for by adjusting the calculated elemental compositions. Any specified chemical modifications are then applied to the heavy and/or light chains. Depending on the type of analysis and the disulfide-bond patterns specified by the user, the number of hydrogens is adjusted for the two polypeptide chains. The glycosylated HC and light chain (LC) (optional) masses are calculated based on the user's input. Finally, multiple HC and LC masses are combined, and the disulfide bond numbers are automatically updated for the intact mass calculation.
With larger molecules such as intact proteins, monoisotopic masses cannot be measured due to the additive mass defect when using mass spectrometers with typical resolving power. Instead, nominal or average masses are measured or reported5,10,11,12,13. The average elemental masses can vary based on the source used for the curated masses14,15. While the differences in elemental masses may be small, they can add up to significant values for large-molecule molecular weight calculations. The average elemental masses used by default in the software application are shown in Supplementary Table 1. For regulated environments like the biopharmaceutical research and development (R&D) field, it is important to maintain consistent molecular masses because changes in masses may imply changes to the molecular entity during regulatory filings. To enable consistency in the use of elemental masses, a dictionary of elemental masses is included with the software tool as a comma-separated value (csv) text file: Element_Mass.csv (Supplementary Coding File 1). Similarly, a curated list of glycan compositions typically seen on mAbs is included: Glycan.csv (Supplementary Coding File 2). Both files are saved in the same folder location as an executable application and can be modified by the user to use a specific elemental mass list or glycan library.
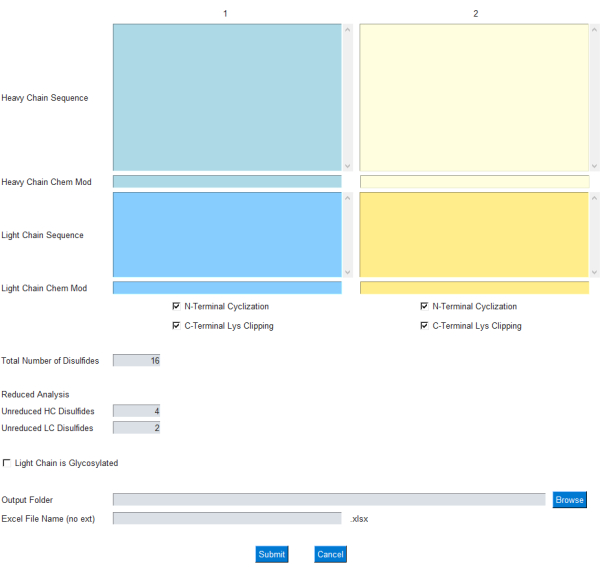
Figure 1: GUI interface for the mAbScale application. The GUI module allows the user to specify the input for the mass calculation. The user enters single-letter amino acid sequences for the light and heavy antibody chains. Common modifications for the heavy-chain N-terminal cyclization and C-terminal lysine clipping are included as check boxes. Chemical formulas/elemental compositions can be added/subtracted through the respective Chem Mod text box. Please click here to view a larger version of this figure.
Protocol
The high-level workflow for mAbScale is shown in Figure 2. Each step has more sophisticated inner decision branches, loops, and combinatorics. A detailed algorithmic workflow describing the calculation process is presented in Supplementary Figure 1. The application output is saved in a spreadsheet format in the user-selected folder. The output file consists of multiple separate worksheets, which can be categorized as the user input, molecular weight calculations, and references for the average isotopic mass derivations (example output is provided in supplemental tables). The user input worksheets include the protein amino acid sequences and other information entered by the user, averaged elemental masses, and glycan masses, which are used to calculate the elemental composition and different molecular weights. The molecular weight calculation sheets include the chemical composition of various forms, the reduced mass with and without glycosylation and chemical modification, and the intact mass with and without glycosylation and chemical modification. Sheets containing half-antibody masses will be generated automatically if the user enters two different HCs and/or two different LCs in the user input page, since half-antibodies are primary impurities that need to be identified and quantified relative to the desired heterodimer. The source code for mAbScale can be accessed through the following repository: https://github.com/kkhatri99/mAbScale.
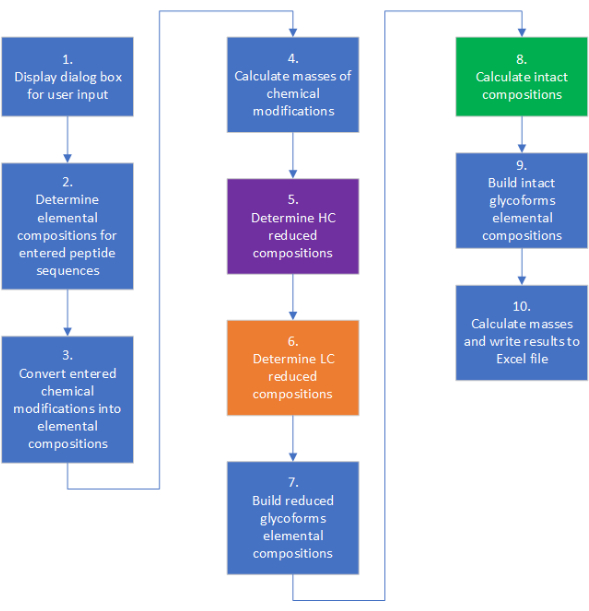
Figure 2: Overview of the steps involved in the calculation of elemental compositions and masses using the application. Color coding can be used to link to the process flow described in Supplementary Figure 1. Please click here to view a larger version of this figure.
1. Opening the mAbscale application
- Open the software application by double-clicking on the icon for the executable file.
2. Sequence entry
- Enter the heavy-chain and light-chain sequences in the respective text boxes marked with 1 without any spaces.
- For bsAbs, add additional heavy or light chains in the second set of text boxes marked 2. Leave 2 blank for mAbs with identical heavy chains and light chains.
- Check the N-Terminal Cyclization and/or C-Terminal Clipping check boxes, if these heavy chain terminal variants are applicable.
- Add any chemical modifications, including linker and payload for ADC molecules, to the Heavy Chain Chem Mod and/or Light Chain Chem Mod text boxes.
- Specify modifications as elemental compositions, such as CaCl2. The modification will be added to the respective protein subunit or chain.
NOTE: A chemical composition may also be subtracted from a subunit or chain by prefixing the elemental composition with a - sign. For example, -H2O will subtract a water molecule from the subunit composition and mass.
- Specify modifications as elemental compositions, such as CaCl2. The modification will be added to the respective protein subunit or chain.
3. Specifying the number of disulfide bonds
- Specify the number of disulfide bonds in the protein molecules in the text box marked Total Number of Disulfides.
- Enter the number of unreduced HC disulfides into the Unreduced HC Disulfides text box and the number of unreduced LC disulfides into the Unreduced LC Disulfides text box, depending on the extent of reduction (complete vs. partial).
NOTE: The reduced mass analysis of mAb subunits involves the reduction/separation of the disulfide-linked heavy and light chains. - If glycosylation is present on the mAb light chain, check the Light Chain is Glycosylated check box.
4. Setting the output folder and running the application
- Click on the Browse button to select an output folder for the Output Folder text box.
- Enter the output file name without a file extension (automatically saves as .xlsx) into the Excel File (no ext) text box.
- Click on the Submit button to start the application. The output file can be found in the designated folder.
NOTE: The elemental masses and the list of glycans can be customized by editing the delimited text files Element_Mass.csv (Supplementary Coding File 1) and Glycan.csv (Supplementary Coding File 2), respectively. These files must be placed in the same folder as the mAbScale.exe (Supplementary Coding File 3) executable file for the application to execute. The application will be closed automatically after one execution. The user will have to start the app again if a second calculation is needed.
Results
A variety of mAbs were selected to represent different types of mAbs. A commercially available mAb standard was selected to represent a conventional mAb with identical heavy chains, identical light chains, and one N-linked glycosylation site in the Fc region. A mAb with an additional light chain N-linked glycosylation, a bispecific mAb, and an antibody-drug conjugate (ADC) mAb were also chosen to widen the application usage. The chemical composition, calculated mass, measured mass, and mass error of these example mA...
Discussion
mAbScale provides an intuitive user interface with the flexibility to alter the building blocks for mass and elemental calculations. The users are expected to have a basic understanding of the target molecule to use the application, derive correct masses, and interpret the results. For example, the intact or reduced mass output sheet can be overwhelming due to the numerous rows of intact or reduced masses, since the default glycan database contains 88 N-linked glycans that are commonly found in the Fc portion of therapeu...
Disclosures
This software is being released under the Apache 2.0 license. Copyright (2022) of GlaxoSmithKline Research & Development Limited. All rights reserved. Licensed under the Apache License, Version 2.0 (the "License"); you may not use this file except in compliance with the License. You may obtain a copy of the License at http://www.apache.org/licenses/LICENSE-2.0. Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "as is" basis, without warranties or conditions of any kind, either expressed or implied. See the License for the specific language governing permissions and limitations under the License. L.C. is a GlaxoSmithKline (GSK) employee. T.H. and K.K. developed this software as employees of GSK and are now associates of Merck and Moderna, respectively.
Acknowledgements
The authors thank Robert Schuster for assistance with data verification.
Materials
| Name | Company | Catalog Number | Comments |
| Acquity UPLC system | Waters Corp., Milford, MA | N/A | Modular system |
| Antibody-drug conjugate (ADC) | GlaxoSmithKline | N/A | Proprietory molecule |
| BEH 200 SEC column | Waters Corp., Milford, MA | 176003904 | |
| Bispecific mAb | GlaxoSmithKline | N/A | Proprietory molecule |
| Byos | Protein Metrics, Cupertino, CA | https://proteinmetrics.com/byos/ Version 4.5 | |
| GPMAW | GPMAW | http://www.gpmaw.com/ | |
| LC-MS grade water | Thermo Fisher Scientific, Waltham, MA | W6-1 | |
| mAb standard | Waters Corp., Milford, MA | 186009125 | Waters Humanized mAb Mass Check Standard |
| mAbScale | GlaxoSmithKline | Apache License, Version 2.0 | |
| Xevo G2 Q-TOF mass spectrometer | Waters Corp., Milford, MA | N/A | Modular system |
References
- Reichert, J. M., Valge-Archer, V. E. Development trends for monoclonal antibody cancer therapeutics. Nature Reviews Drug Discovery. 6 (5), 349-356 (2007).
- Kintzing, J. R., Filsinger Interrante, M. V., Cochran, J. R. Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends in Pharmacological Sciences. 37 (12), 993-1008 (2016).
- Wang, M. -. Y., et al. SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Frontiers in Cellular and Infection Microbiology. 10, 587269 (2020).
- ICH Q8 (R2) Pharmaceutical Development - Scientific Guideline. European Medicines Agency Available from: https://www.ema.europa.eu/en/ch-q8-r2-pharmaceutical-development-scientific-guideline (2018)
- Donnelly, D. P., et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nature Methods. 16 (7), 587-594 (2019).
- Gadgil, H. S., Pipes, G. D., Dillon, T. M., Treuheit, M. J., Bondarenko, P. V. Improving mass accuracy of high performance liquid chromatography/electrospray ionization time-of-flight mass spectrometry of intact antibodies. Journal of the American Society for Mass Spectrometry. 17 (6), 867-872 (2006).
- Beck, A., Sanglier-Cianférani, S., Van Dorsselaer, A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Analytical Chemistry. 84 (11), 4637-4646 (2012).
- Camperi, J., Goyon, A., Guillarme, D., Zhang, K., Stella, C. Multi-dimensional LC-MS: the next generation characterization of antibody-based therapeutics by unified online bottom-up, middle-up and intact approaches. Analyst. 146 (3), 747-769 (2021).
- Liu, H., May, K. Disulfide bond structures of IgG molecules. mAbs. 4 (1), 17-23 (2012).
- Jakes, C., Füssl, F., Zaborowska, I., Bones, J. Rapid analysis of biotherapeutics using protein a chromatography coupled to orbitrap mass spectrometry. Analytical Chemistry. 93 (40), 13505-13512 (2021).
- Robotham, A. C., Kelly, J. F., Matte, A. Chapter 1 - LC-MS characterization of antibody-based therapeutics: Recent highlights and future prospects. Approaches to the Purification, Analysis and Characterization of Antibody-Based Therapeutics. , 1-33 (2020).
- Valeja, S. G., et al. Unit mass baseline resolution for an intact 148 kDa therapeutic monoclonal antibody by fourier transform ion cyclotron resonance mass spectrometry. Analytical Chemistry. 83 (22), 8391-8395 (2011).
- Fornelli, L., Ayoub, D., Aizikov, K., Beck, A., Tsybin, Y. O. Middle-down analysis of monoclonal antibodies with electron transfer dissociation orbitrap fourier transform mass spectrometry. Analytical Chemistry. 86 (6), 3005-3012 (2014).
- Berglund, M., Wieser, M. E. Isotopic compositions of the elements 2009 (IUPAC Technical Report). Pure and Applied Chemistry. 83 (2), 397-410 (2011).
- Wang, M., et al. The Ame2012 atomic mass evaluation. Chinese Physics C. 36 (12), 1603-2014 (2012).
- Peri, S., Steen, H., Pandey, A. GPMAW--A software tool for analyzing proteins and peptides. Trends in Biochemical Sciences. 26 (11), 687-689 (2001).
- Tipton, J. D., et al. Analysis of intact protein isoforms by mass spectrometry. The Journal of Biological Chemistry. 286 (29), 25451-25458 (2011).
- De Leoz, M. L. A., et al. interlaboratory study on glycosylation analysis of monoclonal antibodies: Comparison of results from diverse analytical methods. Molecular & Cellular Proteomics. 19 (1), 11-30 (2020).
- Cymer, F., Beck, H., Rohde, A., Reusch, D. Therapeutic monoclonal antibody N-glycosylation - Structure, function and therapeutic potential. Biologicals. 52, 1-11 (2018).
- Baker, P. R., Trinidad, J. C., Chalkley, R. J. Modification site localization scoring integrated into a search engine. Molecular & Cellular Proteomics. 10 (7), (2011).
- Chalkley, R. J., Clauser, K. R. Modification site localization scoring: Strategies and performance. Molecular & Cellular Proteomics. 11 (5), 3-14 (2012).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionExplore More Articles
This article has been published
Video Coming Soon
ISSN 2578-2037
Copyright © 2025 MyJoVE Corporation. All rights reserved
We use cookies to enhance your experience on our website.
By continuing to use our website or clicking “Continue”, you are agreeing to accept our cookies.