A subscription to JoVE is required to view this content. Sign in or start your free trial.
Method Article
An Acetyl-Click Chemistry Assay to Measure Histone Acetyltransferase 1 Acetylation
In This Article
Summary
Quick and accurate chemical assays to screen for specific inhibitors are an important tool in the drug development arsenal. Here, we present a scalable acetyl-click chemistry assay to measure the inhibition of HAT1 acetylation activity.
Abstract
HAT1, also known as Histone acetyltransferase 1, plays a crucial role in chromatin synthesis by stabilizing and acetylating nascent H4 before nucleosome assembly. It is required for tumor growth in various systems, making it a potential target for cancer treatment. To facilitate the identification of compounds that can inhibit HAT1 enzymatic activity, we have devised an acetyl-click assay for rapid screening. In this simple assay, we employ recombinant HAT1/Rbap46, which is purified from activated human cells. The method utilizes the acetyl-CoA analog 4-pentynoyl-CoA (4P) in a click-chemistry approach. This involves the enzymatic transfer of an alkyne handle through a HAT1-dependent acylation reaction to a biotinylated H4 N-terminal peptide. The captured peptide is then immobilized on neutravidin plates, followed by click-chemistry functionalization with biotin-azide. Subsequently, streptavidin-peroxidase recruitment is employed to oxidize amplex red, resulting in a quantitative fluorescent output. By introducing chemical inhibitors during the acylation reaction, we can quantify enzymatic inhibition based on a reduction of the fluorescence signal. Importantly, this reaction is scalable, allowing for high throughput screening of potential inhibitors for HAT1 enzymatic activity.
Introduction
Among the numerous eukaryotic acetyltransferases, HAT1 was the initial histone acetyltransferase to be isolated1,2,3. Subsequent investigations have firmly established its pivotal role in chromatin replication, particularly in the synthesis of new nucleosomes during S-phase4. Our research endeavors led to the recognition that HAT1 is highly stimulated by epidermal growth factor (EGF) treatment in mammary cells5. Furthermore, it has come to light that HAT1 is required for rapid cell proliferation and tumor formation in vivo6,7,8,9. Data indicate that HAT1 is critical in coordinating anabolic and epigenetic processes for cell division, driving tumor growth.
HAT1 di-acetylates the amino-terminal tail of histone H4 on lysines 5 and 12 in complex with the chaperone protein Rbap46, which binds the histone and presents the amino-terminus to HAT1. Histone tetramers or disomes10, together with HAT1/Rbap46 and other histone chaperones11, are then imported to the nucleus. Histones are then released to be deposited at the replication fork, or other sites to support gene activation or repression. The function of the HAT1 di-acetylation mark on histone H4 is not fully understood. It is likely quickly removed within a span of 15-30 min by the action of histone deacetylases12,13,14,15 after H4 is inserted into chromatin. Thus, the HAT1 di-acetylation mark is not propagated in chromatin and may not serve a true epigenetic role, although a role in the recruitment of chromatin-modifying enzymes to nascent chromatin has been postulated12. Also, HAT1 does not directly acetylate chromatin; its activity is restricted to soluble histones.
The development of small-molecule histone acetyltransferase inhibitors has been hampered by nonspecific and low-throughput assays, often resulting in the generation of biologically reactive compounds16,17. The gold-standard assay to measure acetyltransferase activities requires the use of 3H-acetyl-coA, which limits throughput and requires radiation. Nonetheless, recently, specific and highly potent small molecule acetyltransferase inhibitors targeting CBP/p30018 and KAT6A/B19,20 have been described and confirmed through the use of 3H-acetyl-CoA. Moving forward, improved assays to achieve better throughput and avoid laboratory hazards are being devised.
Recent advances in acetylation monitoring21 have used click chemistry to enable enzymatic reaction monitoring. There are a variety of click-enabled precursors that are accessible by simple synthetic routes or available for purchase that can be incorporated into enzyme reactions. These reactions are typically carried out in recombinant systems, although cell-based assays are also feasible22. The advantage of click-enabled co-factors and substrates is that screening can directly measure enzyme activity without the need for coupled read-out systems that are often perturbed by screening compounds and require additional handling steps. This allows for inhibitor treatments only during the enzymatic step, whereas all downstream functionalization and detection steps are carried out after extensive washing to remove compounds, thus limiting the potential for assay interference to occur. These advantages make the design of click-enabled assays preferable to coupled assays that commonly rely on the detection of free coenzyme A.
One important consideration is the acceptance of click-enabled co-factors into the enzyme active site. Existing click-enabled co-factors may not be fully compatible with the active site optimized for the native co-factor. Structural information and modeling can be used to design amino acid substitutions to enlarge the active site to incorporate altered substrates23. This may enable screening with improved enzyme kinetics and lower substrate and enzyme levels. The drawback of this approach is that altered catalytic pockets may not identify inhibitors that interact strongly with the native enzyme. Ultimately, a combination of approaches is required to identify and validate potential enzyme inhibitors.
Here, we describe a method developed to purify and assay HAT1 enzyme activity using the click co-factor 4-pentynoyl-CoA24. This assay (Figure 1) uses the native enzyme sequence in complex with its required partner protein Rbap46, which has been shown to boost enzyme activity. Purification of the enzyme from human cells allows for enzyme activation in cellulo, which may preserve stimulating post-translational modifications important for full enzyme activity. Design and optimization of recombinant enzyme assays for high-throughput chemical screens have successfully been used to identify and characterize HAT1 small molecule inhibitors.
Protocol
1. Method 1: Producing and purifying recombinant HAT1/Rbap46 complex
- Thawing, recovering, and expanding HEK293f cells
- Thaw 1-10 million HEK293f mammalian cells26 into 30 mL freestyle 293 expression media in a 100 mL flask. Incubate in 8% CO2 at 37 °C while rotating at 60 rpm.
- The next day, count and check cell viability, then adjust rotation speed to 120 rpm. Expand to 300 mL culture in a 1 L flask, maintaining seeding density at 500,000 cells/mL and splitting cells before density exceeds 3 x 106 cells/mL.
- HEK293f transfection
- Seed 5 x 105 cells/mL in 300 mL culture in a 1 L flask and culture for 24 h. The next day, count cells to ensure they are between 7.5 x 105 to 1.2 x 106 cells/mL.
- Prepare a mixture of 300 µg of pHEK-FLAG-HAT1 and 300 µg of pHEK-Rbap46 plasmid DNAs in 30 mL of PBS. Add 1.2 mL of polyethylenimine (PEI) to the DNA/PBS and mix. Incubate for 20 min at RT. Add the DNA/PBS/PEI mix to the 300 mL culture and incubate at 37 °C for 48 h at 120 rpm.
- Harvesting cells
- Count cells to ensure they fall between 1.7 x 106 to 2.5 x 106 cells/mL. At this cell density, add forskolin to a final concentration of 12.5 µM to activate HAT1 and incubate cells for 30 min, 37 °C, 120 rpm.
- Pellet the cells at 300 x g for 5 min, wash once with 30 mL of PBS, and snap-freeze in liquid N2. Store at -80 °C until protein purification or proceed directly to purification.
- HAT1/Rbap46 complex FLAG-bead purification
- Lyse cells and prepare protein extract.
- Prepare lysis buffer by adding one tablet of protease inhibitor cocktail (PIC) to 50 mL of RSB-500 (20 mM Tris-HCl pH 7.5, 500 mM NaCl, 25 mM MgCl2).
- Thaw the cell pellet from 300 mL culture on ice and lyse with 40 mL of ice-cold lysis buffer with 0.1% Triton X-100. Sonicate on ice, then spin down at 10,000 x g for 10 min at 4°C. Collect all supernatant into one 50 mL tube on ice, which now is the protein extract.
- Prepare FLAG beads.
- Split the FLAG beads into four 15 mL conical tubes by adding 400 µL of M2-FLAG agarose bead slurry into each tube containing 5 mL of 0.1 M Glycine pH 3.5, 0.01% Triton X-100. Shake each tube vigorously for 5 s by hand.
- Pulse centrifuge for 30 s at 1000 x g at 4 °C, aspirate supernatant, wash twice with 5 mL of RSB-500 + 0.1% Triton X-100 (Tx-100; no protease inhibitor), and incubate on ice.
- Perform FLAG immunoprecipitation.
- Add 10 mL of protein extract to each 15 mL conical tube containing washed beads. Incubate the tubes at 4 °C with inverted rotation for at least 90 min or overnight.
- Wash the bound beads 5x in 10 mL of RSB-500 + 0.1% Tx-100. For each wash, invert the tube 2x-5x to resuspend beads, then pellet at 1000 x g for 1 min at 4 °C. Finally, wash 1x in RSB-100 (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 25 mM MgCl2) + 0.1% Tx-100.
- FLAG peptide elution: Elute the HAT1 complex in 1.5 mL of Elution Buffer (EB) containing 0.5 mg/mL FLAG peptide. Incubate at 4 °C for at least 1 h or overnight. Pulse centrifuge the eluted beads at 1000 x g for 30 s at 4 °C and collect the supernatant, which contains the purified HAT1/Rbap46 complex.
- Concentrate protein and remove FLAG peptide.
- Add eluate to a 20 mL, 10,000 Dalton cutoff filter tube. Bring the volume up to 15 mL with EB. Spin at 2500 x g for 15 min at 4 °C, repeating twice with an additional 15 mL of EB each time.
- Recover the final eluate from the filter tube and check the protein concentration by A260. Expect a concentration of 1 mg total protein in 6 mL of EB (per 300 mL starting culture). Check protein purity by SDS-PAGE, Coomassie, and immunoblotting, and store aliquots at -80 °C.
- Lyse cells and prepare protein extract.
2. Method 2: HAT1 acetyl-click standard curve
- Preparing standard curve
- Synthesize a positive control H4 N-terminal peptide with the sequence: SCRG[Pra]GGKGLG[Pra]GGAKRHRKVLRGG[Lys(Biotin)], where [Pra] denotes Propargylglycine.
- Resuspend the Pra peptide to 0.1 mg/mL in DMSO. Mix the Pra peptide with biotinylated histone H4 peptide (1-23-GGK-biotin) to create a standard curve (Figure 2; volumes found in Table 1). Standard curves may be made fresh prior to plate binding or, if made in advance, mixed with 20 µL of 8 M urea and stored at -20 °C until step 3.3.
3. Method 3: HAT1 acetyl-click assay
- Assembling acetylation reactions with test inhibitors
- Assemble acetylation reactions in duplicate in 0.2 mL PCR tubes or 96-well PCR plates from the following components: biotinylated histone H4 peptide (1-23-GGK-biotin) resuspended in DMSO to 0.1 mg/mL (34.8 µM), HAT1 enzyme pre-diluted in EB, 20x buffer (1M Tris pH 8.5, 0.1% NP40), 2 mM DTT, 4-pentynoyl-CoA dissolved in water to 1 mg/mL (1 mM). A 20 µL reaction comprises 10 µL of enzyme, 1 µL of H4 peptide, 1 µL of 20x buffer, 1 µL of DTT pre-mixed and aliquoted to wells of a 96-well PCR plate on ice.
- Add 1 µL of DMSO (negative control) or test compound dissolved at 1-10 µM, or H4K12CoA25 (or suitable positive control inhibitor), per well. Mix by gentle pipetting and incubate for 10 min on ice to allow enzyme: inhibitor complexes to form.
- Acetylation reaction continuation
- Combine 2 µL of 4-pentynoyl-CoA with 4 µL of water, then add to the wells. Gently mix by pipetting, centrifuge at 300 x g for 30 s at RT to collect the contents, and incubate at 37 °C for 1 h in capped tubes or plates with resealable foil.
- Process the contents directly for reaction products or quench with 20 µL of 8 M urea and store at -20 °C until processing.
- Peptide binding to Neutravidin plate
- Add reaction contents with or without urea to bovine serum albumin (BSA) pre-blocked black Neutravidin-coated 96-well plates containing 80 µL of PBST (PBS + 0.1% Tween-20) per well.
- Add standard curve peptides in duplicate to their own wells containing 80 µL of PBST. Bind peptides with gentle orbital shaking for 1 h at room temperature (RT).
- Wash: After binding is complete, aspirate the liquid from the wells and wash the wells with 200 µL of PBST 3x 15 strokes (180 µL stroke volume) using a plate washer.
- Click chemistry reaction
- Prepare the reagents for the click reaction as follows: 100 mM Tris(3-hydroxypropyltriazolymethyl)amine (THPTA) ligand in water and 20 mM CuSO4 in water. THPTA prevents Cu(II) catalyzed hydrolysis and quenches radicals and peroxides generated from O2/Cu/ascorbate.
- Add the THPTA/Cu mixture to 300 mM sodium ascorbate in water and 2.5 mM biotin azide in DMSO. One-click reaction contains 140 µL of PBS, 10 µL of THPTA, 10 µL of CuSO4, 10 µL of sodium ascorbate, 20 µL of biotin-azide. Always mix the click reagents fresh, then dispense 190 µL to each well, seal the plate, and incubate at 37°C for 1 h.
- Wash: Aspirate the liquid from the wells and wash the plate 3x with PBST (180 µL stroke volume, 15 strokes for each wash).
- Streptavidin-horseradish peroxidase binding: Dilute streptavidin-HRP (0.224 mg/mL) 1:10 in streptavidin (0.224 mg/mL), then further dilute 1:1000 in PBST. Add Steptavidin-HRP: streptavidin mix (100 µL per well) and incubate at RT for 1 h with gentle orbital shaking.
- Wash 3x with PBST as in the previous steps.
- Amplex red oxidation
- Combine the Amplex red detection reagents as follows: 4.45 mL of NaHPO4 buffer (1x = 50 mM final concentration), 50 µL amplex red (20 mM diluted in DMSO), 500 µL of diluted H2O2.
- Dilute H2O2 from 30% stock to 3% in 1x NaHPO4 buffer, then add 22.7 µL of 3% H2O2 into 977 µL of 1x NaHPO4 buffer (this is the H2O2 used for the amplex reaction). Add 100 µL of the amplex red reaction mixture per well, incubate at RT for 30 min protected from light, then detect the fluorescence excitation/emission 571/585 nm using a standard fluorescence plate reader.
- Calculation of enzyme inhibition: Calculate percent inhibition (Figure 3) according to the following formula:
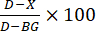 , where D is the fluorescence value of control reactions treated with DMSO only, X is the value of reactions treated with test compounds, and BG is the value of background wells (H4 peptide control, no enzyme added).
, where D is the fluorescence value of control reactions treated with DMSO only, X is the value of reactions treated with test compounds, and BG is the value of background wells (H4 peptide control, no enzyme added). - Dose curve: Select test compounds that are found to inhibit HAT1 enzyme activity for repeat assays, with the compound serially diluted. Determine the compound dilutions empirically. For example, start with 2 mM stock concentration, then serially dilute 1:3 for eight dilutions. Then dilute 1:20 into enzyme assays yielding 100 µM top dose.
- Plotting the curve: Use least squares regression to fit dose-response inhibition curves (using percent inhibition values from step 3.10) in data analysis software (GraphPad Prism) and derive IC50 values for each compound (Figure 4).
- Acetylation reaction with acetyl-CoA:
- Use this to confirm enzyme activity with the native co-factor acetyl-CoA instead of 4-pentynoyl-CoA. Carry out HAT1 acetylation assays as described in steps 3.1-3.2 with acetyl-CoA in place of 4-pentynoyl-CoA.
- Spot the reaction products onto nitrocellulose membranes (1-2 µL), allow them to dry, then dot-blot with anti-H4-lysine-12-acetyl or anti-H4-lysine-5-acetyl antibodies. Quantify the immunoblot signal by densitometry.
Results
Standard curves in duplicate (16 wells) should be included on every plate to ensure proper assay performance. Standard curve data should be set up in table form, with a range of 100% to 0% according to the ratio of Pra-containing peptide to native H4 peptide in solution (Table 1). Amplex red signal will be the highest in 100% pra/0% native H4 peptide wells, and lowest in 0% pra/100% native H4 peptide wells. After fluorescence has been detected and wells are averaged, the resulting standards graph should ...
Discussion
In the past decade, click chemistry became prominent20, enabling the precise design of interacting chemical structures. Within this context, various bioorthogonal covalent connections21 have emerged as promising options for forming complexes in their natural environment. Click chemistry employs pairs of functional groups that exhibit rapid and selective reactions, commonly known as "click reactions." These reactions occur efficiently in environmentally friendly, gen...
Disclosures
We have filed patents describing the HAT1 acetyl-click assay (PCT/US20/29395). J.J.G. reports consulting agreements with Sharma Therapeutics, LLC and Guidepoint and research support (to his institution) from Hummingbird Biosciences.
Acknowledgements
We thank George Zheng for providing H4K12CoA. We thank members of the Gruber Lab for helpful discussions and feedback. We thank support from the NIH/NCI (1K08CA245024), CPRIT (RR200090, RP210041), and the V Foundation (V2022-022).
Materials
| Name | Company | Catalog Number | Comments |
| 4P CoA | Cayman Chemical | 10547 | Click chemistry co-factor |
| Amplex Red | Fisher Sci | A12222 | Fluorescence substrate |
| Biotin-PEG-Azide | Alfa Aesar | J64996MC | Click chemistry |
| Copper Sulfate | Sigma-aldrich | 7758-98-7 | Click chemistry |
| DMSO | Fisher Scientific | 67-68-5 | diluent |
| DTT | Acros Organics | 03-12-3483 | reducting agent |
| Forskolin | VWR | 102987-310 | Protein expression |
| Freestyle 293 Expression Medium | Thermo Fisher | 12338018 | Media |
| Freestyle 293-F cells | Thermo Fisher | R790-07 | Protein expression |
| H4-peptide/1-23-GGK-biotin | Anaspec | AS65097 | peptide substrate |
| HEPES | Sigma-aldrich | 7365-45-9 | EB buffer |
| Hydrogen peroxide 30% solution | Sigma-aldrich | Z00183-99-0 | initiator |
| M2 FLAG antibody slurry | Millipore-Sigma | A2220 | Protein purification |
| Macrosep 10K Filter (Pall Lab) | VWR | 89131-980 | Protein purification |
| Neutravidin Plate | Thermo Sci | 15127 | BSA-pre-blocked |
| NP40 (IGEPAL) | MP Biomedical | 198596 | 20x buffer |
| pHEK-293 plasmid | Takara Bio | 3390 | Protein expression |
| Phosphate Buffered Saline 10x | Alfa Aesar | Z00082-33-6 | wash buffer |
| Pra peptide | Genscript | Custom synthesis | biotinylated |
| Sodium Ascorbate | Sigma-aldrich | 134-03-2 | Click chemistry |
| Sodium chloride | Sigma-aldrich | 7647-14-5 | EB buffer |
| Sodium phosphate | VWR International | 7558-80-7 | buffer |
| Streptavidin | EMD Millipore | 189730 | competitor |
| Streptavidin-HRP | Cell Signaling | 3999S | enzyme |
| THPTA ligand | Fisher Sci | 1010-500 | Click chemistry |
| Tris base | Sigma-aldrich | 77-86-1 | 20x buffer |
| Triton-X 100 | VWR International | 9002-93-1 | EB buffer |
| Tween-20 | Sigma-aldrich | 9005-64-5 | Wash buffer |
| Urea | Sigma-Aldrich | 57-13-6 | quencher |
References
- Kleff, S., et al. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 270 (42), 24674-24677 (1995).
- Parthun, M. R., Widom, J., Gottschling, D. E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 87 (1), 85-94 (1996).
- Verreault, A., et al. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 8 (2), 96-108 (1998).
- Parthun, M. R. Histone acetyltransferase 1: more than just an enzyme. Biochim Biophys Acta. 1819 (3-4), 256-263 (2013).
- Gruber, J. J., et al. HAT1 coordinates histone production and acetylation via H4 promoter binding. Mol Cell. 75 (4), 711-724 e5 (2019).
- Fan, P., et al. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J Exp Clin Cancer Res. 38 (1), 47 (2019).
- Xia, P., et al. MicroRNA-377 exerts a potent suppressive role in osteosarcoma through the involvement of the histone acetyltransferase 1-mediated Wnt axis. J Cell Physiol. 234 (12), 22787-22798 (2019).
- Yang, G., et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 22 (2), e50967 (2021).
- Xue, L., et al. RNAi screening identifies HAT1 as a potential drug target in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 7 (7), 3898-3907 (2014).
- Zhang, W., et al. Structural plasticity of histones H3-H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat Struct Mol Biol. 20 (1), 29-35 (2013).
- Campos, E. I., et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 17 (11), 1343-1351 (2010).
- Agudelo Garcia, P. A., et al. Identification of multiple roles for histone acetyltransferase 1 in replication-coupled chromatin assembly. Nucleic Acids Res. 45 (16), 9319-9335 (2017).
- Nagarajan, P., et al. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 9 (6), e1003518 (2013).
- Annunziato, A. T. Assembling chromatin: the long and winding road. Biochim Biophys Acta. 1819 (3-4), 196-210 (2013).
- Annunziato, A. T., Seale, R. L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 258 (20), 12675-12684 (1983).
- Dahlin, J. L., et al. Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nat Commun. 8 (1), 1527 (2017).
- Baell, J. B., Miao, W. Histone acetyltransferase inhibitors: where art thou. Future Med Chem. 8 (13), 1525-1528 (2016).
- Lasko, L. M., et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 550 (7674), 128-132 (2017).
- Baell, J. B., et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 560 (7717), 253-257 (2018).
- Falk, H., et al. An efficient high-throughput screening method for MYST family acetyltransferases, a new class of epigenetic drug targets. J Biomol Screen. 16 (10), 1196-1205 (2011).
- He, M., et al. Chemical biology approaches for investigating the functions of lysine acetyltransferases. Angew Chem Int Ed Engl. 57 (5), 1162-1184 (2018).
- Lipchik, A. M., et al. A peptide-based biosensor assay to detect intracellular Syk kinase activation and inhibition. Biochemistry. 51 (38), 7515-7524 (2012).
- Song, J., et al. Chemoproteomic profiling of protein substrates of a major lysine acetyltransferase in the native cellular context. ACS Chem Biol. 17 (5), 1092-1102 (2022).
- Gaddameedi, J. D., et al. Acetyl-click screening platform identifies small-molecule inhibitors of histone acetyltransferase 1 (HAT1). J Med Chem. 66 (8), 5774-5801 (2023).
- Ngo, L., Brown, T., Zheng, Y. G. Bisubstrate inhibitors to target histone acetyltransferase 1 (HAT1). Chem Biol Drug Des. 93 (5), 865-873 (2019).
- Parker, C. G., Pratt, M. R. Click chemistry in proteomic investigations. Cell. 180 (4), 605-632 (2020).
- Islam, K. The bump-and-hole tactic: Expanding the scope of chemical genetics. Cell Chem Biol. 25 (10), 1171-1184 (2018).
- Radziwon, K., Weeks, A. M. Protein engineering for selective proteomics. Curr Opin Chem Biol. 60, 10-19 (2021).
- Rich, R. L., Myszka, D. G. Survey of the year 2007 commercial optical biosensor literature. J Mol Recognit. 21 (6), 355-400 (2008).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved