A subscription to JoVE is required to view this content. Sign in or start your free trial.
Methods Article
Rapid Detection of Bacterial Pathogens Causing Lower Respiratory Tract Infections via Microfluidic-Chip-Based Loop-Mediated Isothermal Amplification
* These authors contributed equally
In This Article
Summary
Various bacterial pathogens can cause respiratory tract infections and lead to serious health issues if not detected accurately and treated promptly. Rapid and accurate detection of these pathogens via loop-mediated isothermal amplification provides effective management and control of respiratory tract infections in clinical settings.
Abstract
Respiratory tract infections (RTIs) are among the most common problems in clinical settings. Rapid and accurate identification of bacterial pathogens will provide practical guidelines for managing and treating RTIs. This study describes a method for rapidly detecting bacterial pathogens that cause respiratory tract infections via multi-channel loop-mediated isothermal amplification (LAMP). LAMP is a sensitive and specific diagnostic tool that rapidly detects bacterial nucleic acids with high accuracy and reliability. The proposed method offers a significant advantage over traditional bacterial culturing methods, which are time-consuming and often require greater sensitivity for detecting low levels of bacterial nucleic acids. This article presents representative results of K. pneumoniae infection and its multiple co-infections using LAMP to detect samples (sputum, bronchial lavage fluid, and alveolar lavage fluid) from the lower respiratory tract. In summary, the multi-channel LAMP method provides a rapid and efficient means of identifying single and multiple bacterial pathogens in clinical samples, which can help prevent the spread of bacterial pathogens and aid in the appropriate treatment of RTIs.
Introduction
Respiratory tract infections (RTIs) caused by bacterial pathogens primarily contribute to morbidity and mortality worldwide1. It is defined as any upper or lower respiratory symptoms accompanied by fever lasting 2-3 days. While upper respiratory infection is more common than lower respiratory infection, chronic and recurrent respiratory tract infections are also common clinical conditions, posing great risks to individuals and placing a significant burden on healthcare systems2. Common bacterial pathogens of RTIs include Streptococcus pneumoniae3, Haemophilus influenzae4, Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Stenotrophomonas maltophilia, among others. These pathogenic bacteria usually colonize the mucosal surfaces of the host's nasopharynx and upper respiratory tract, causing typical symptoms of RTIs such as sore throat and bronchitis. They cause pneumonia when they spread from the upper respiratory tract to sterile areas of the lower respiratory tract and may spread from person to person through the respiratory tract5. In severe cases, they can also lead to invasive bacterial diseases, especially bacteremic pneumonia, meningitis, and sepsis, which are leading causes of morbidity and mortality in people of all age groups worldwide.
Traditional tests for RTIs involve microbiological culture using throat swabs and sputum respiratory samples6. Additionally, serological tests such as enzyme-linked immunosorbent assay (ELISA) detect antibodies or antigens in serum, while agglutination tests observe the agglutination reaction of antibodies and antigens to detect infection7. Microbial culture is considered the gold standard for diagnosing RTIs, but its low culture positivity rate, poor reliability, and long detection cycle limit diagnostic efficiency8. In reality, rapid and accurate diagnosis of RTIs is crucial for precise eradication of the bacterial pathogen. Quick and effective detection methods can help reduce the transmission rate of pathogens, shorten the duration of infection, and decrease unnecessary antibiotic use9,10. Molecular biology-based methods significantly expedite detection, such as polymerase chain reaction (PCR), which amplifies a target gene's DNA sequence to detect pathogens. However, traditional PCR necessitates complex temperature cycling equipment, which is cumbersome and time-consuming. Furthermore, every DNA amplification using PCR (except real-time PCR) concludes with electrophoretic separation of the product, which also takes time. Visualization of the product requires dyes, many of which are mutagenic or carcinogenic. Therefore, it is imperative to continuously develop new methods and technologies for diagnosing RTI bacterial pathogens.
Loop-Mediated Isothermal Amplification (LAMP) is a novel and emerging molecular technology initially developed by Notomi et al. in 200011. LAMP can amplify DNA under stable isothermal conditions without complex temperature cycling equipment, which makes it suitable for rapid detection and reduces equipment complexity and cost12. LAMP can detect low concentrations of target DNA with high sensitivity13. It uses multiple specific primers to improve selectivity for target sequences and reduce the possibility of false positives14. LAMP is gradually being widely used in clinical laboratories due to its ease, speed, and intuitive operation, even for detecting RTIs. In this study, we investigated the effectiveness of LAMP in detecting lower RTIs in clinical samples (sputum, bronchial lavage fluid, and alveolar lavage fluid), as shown in Figure 1. It is evident that LAMP offers advantages such as speed, sensitivity, and ease of use over traditional tests in lower RTI detection, making it a promising application.
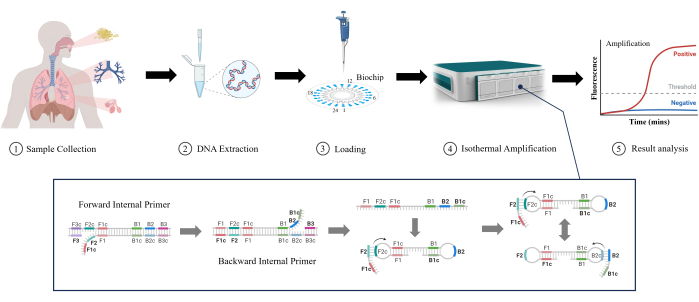
Figure 1: Schematic illustration of the LAMP detection method. Please click here to view a larger version of this figure.
Protocol
All samples for this study were evaluated and approved by the Ethics Review Committee of Guangdong Provincial People's Hospital (Approval Number: KY2023-1114-01). All participants signed written informed consent before the experiments. The reagents and equipment used for the study are listed in the Table of Materials. The abbreviations used in the protocol are listed in Supplementary Table 1.
1. Collection of clinical samples from the lower respiratory tract
- Sputum collection
- Cleanse the oral cavity and teeth with clean water, ensuring denture removal if applicable. Cough forcefully to expel deep respiratory sputum into a sputum container (minimum 0.6 mL).
- Avoid contamination with saliva or nasal discharge. If coughing is difficult, administer inhalation of a 100 g/L NaCl solution at 45 °C to facilitate expectoration. Transfer the specimen to a sterile, sealable container.
NOTE: Ensure the sputum samples are collected in the morning and before any antibacterial drug use.
- Bronchial lavage fluid (BLF)
- Insert the collector head into the trachea from the nostril or tracheostomy site (approximately 30 cm deep). Inject 5 mL of saline, establish negative pressure, rotate the collector head, and slowly withdraw.
- Collect the extracted mucus and rinse the collector once with a sampling solution (physiological saline or sterile water for injection). Alternatively, connect a pediatric urinary catheter to a 50 mL syringe as a substitute for collection.
- Alveolar lavage fluid (ALF)
- Identify the lesion location through chest imaging examinations. Select the most significant or rapidly progressing area for lavage.
- Inject 1-2 mL of 2% lidocaine into the bronchial segment through the biopsy channel during lavage, providing local anesthesia for the lavaged lung segment15. For patients under intravenous general anesthesia who still exhibit strong airway reactions, inject 1-2 mL of 2% lidocaine.
- After local anesthesia, insert a fiberoptic bronchoscope through the mouth or nose, passing through the pharynx into the bronchus of the right middle lobe or the lingual segment of the left lung. Place its tip at the opening of the bronchial branch and slowly introduce sterile physiological saline.
- Administer 30-50 mL saline per session, with 100-250 mL volume (max. 300 mL). Collect the extracted mucus in a sealed sterile container.
2. DNA extraction
- Based on the viscosity of lower respiratory tract samples, add an appropriate amount of 10% NaOH. For samples with low viscosity, add 2-3 times the volume of the sample of liquefying solution. For samples with moderate viscosity, add 5-6 times the volume of the sample of liquefying solution. For samples with high viscosity, add 8-10 times the volume of the sample of liquefying solution.
- Adjust the volume of 10% NaOH according to the sample viscosity. Disperse the samples as uniformly as possible using a vortex mixer for 15 s and incubate at 37 °C for 30 min for liquefaction.
NOTE: For thicker samples, increase the volume of NaOH or extend the liquefaction time. The liquefaction quality of samples directly influences the subsequent extraction efficiency. Ideally, liquefied samples should exhibit a consistent, non-sticky consistency.
- Adjust the volume of 10% NaOH according to the sample viscosity. Disperse the samples as uniformly as possible using a vortex mixer for 15 s and incubate at 37 °C for 30 min for liquefaction.
- Pipette 1 mL of the liquefied sample into a 1.5 mL centrifuge tube. Centrifuge at a speed of 15,777 × g for 5 min at 2-6 °C, then use a pipette to remove and discard the supernatant.
NOTE: When aspirating the sample, avoid drawing impurities from the bottom of the tube; if there are many impurities, the sample can be centrifuged at 1,753-2,739 × g for 1 min before aspiration. - Add 1 mL of washing solution to the centrifuge tube and vortex to lift the precipitate from the bottom of the tube. There is no need to disperse it completely.
- Centrifuge the solution at 15,777 × g for 5 min, discard the supernatant, and try not to touch the precipitate.
NOTE: Ensure thorough removal of the washing solution to avoid affecting subsequent amplification.
- Centrifuge the solution at 15,777 × g for 5 min, discard the supernatant, and try not to touch the precipitate.
- Add 100 µL of nucleic acid extraction solution to the centrifuge tube. Use a pipette to aspirate and mix the precipitate thoroughly. Transfer the liquid and precipitate together into a nucleic acid extraction tube. The composition of the nucleic acid extraction solution is presented in Table 1.
- Place the nucleic acid extraction tubes in a vortex mixer and vortex at medium speed for at least 5 min. After vortexing, transfer the nucleic acid extraction tubes to a metal bath with a constant temperature and heat at 100 °C for 5 min.
- Centrifuge at 15,777 × g at 2-6 °C for 5 min and set aside.
NOTE: If the PCR amplification reaction is performed within 24 h, the nucleic acid can be stored at 4 °C. After the amplification, store the nucleic acid at -20 °C. For long-term storage (beyond 24 h), store the nucleic acid at -20 °C. When ready to use again, thaw the sample naturally, vortex to mix, heat in a 95 °C water bath for 5 min, centrifuge at 10,956 × g for 1 min, and use the supernatant for PCR amplification.
| Solution | Components | Number | Specification |
| Washing Solution | 10 mM EDTA | 1 bottle | 24 mL/bottle |
| Nucleic Acid Extraction Reagent | 10mM Tris-HCl, 1mM EDTA, Nucleic acid preservatives | 2 tubes | 1.2 mL/bottle |
| Nucleic Acid Extraction Tube | Glass beads | 1 bag | 24 bottles/bag |
Table 1: Composition of nucleic acid extraction reagent.
3. Loop-mediated isothermal amplification and microfluid chip
- Reagent and microfluidic chip
- Conduct isothermal amplification reactions on a microfluidic disk-shaped chip (see Table of Materials). Carry out the constant-temperature amplification at 65 °C.
- Perform real-time fluorescence analysis using a fluorescent dye incorporation method14 on the constant-temperature amplification microfluidic chip nucleic acid analyzer. Observe an S-shaped amplification curve for positive samples using a polymerase with strand displacement functionality.
- Loop-mediated isothermal amplification
- Target six sequence regions with four specific primers, including two inner primers and two outer primers (provided with the LAMP kit).
NOTE: DNA is continuously replicated and amplified at a constant temperature using a DNA polymerase with strand displacement functionality. The reaction process involves the dumbbell-shaped template synthesis stage, cycling amplification stage, elongation, and recycling stage, ultimately forming a mixture of DNA fragments with stem-loop and cauliflower-like structures. Refer to the kit instructions for detailed information (Supplementary File 1). - Add two-loop primers to the reaction system to enhance reaction efficiency, which binds to the stem-loop structures, initiating strand displacement synthesis and cyclic replication. The composition of the nucleic acid detection kit for respiratory tract pathogens is presented in Table 2.
NOTE: The reagent kit utilizes the LAMP method. The principle is based on the dynamic equilibrium of DNA at around 65 °C, where any primer extending through base pairing at the complementary site of double-stranded DNA causes the other strand to dissociate, forming a single strand.
- Target six sequence regions with four specific primers, including two inner primers and two outer primers (provided with the LAMP kit).
- Microfluidic chip
NOTE: Each microfluidic chip (see Table of Materials) is equipped with 24 reaction wells numbered counterclockwise, with the inlet and outlet for well 1 corresponding to the reaction well 1 (Figure 2). A specific set of primers for amplification and detection of a particular nucleic acid target sequence is embedded and fixed in each reaction well.- Mix the sample DNA with the amplification reagent. Inject the mixture into the chip and distribute it to each reaction well centrifuge at 1,000 × g for 30 s at room temperature.
NOTE: Independent isothermal amplification reactions and real-time fluorescence detection co-occur in each reaction well on the chip. If an S-shaped amplification curve is detected in a particular reaction well, the corresponding detection index for that well is positive.
- Mix the sample DNA with the amplification reagent. Inject the mixture into the chip and distribute it to each reaction well centrifuge at 1,000 × g for 30 s at room temperature.
| Components | Composition | Number |
| Chip | Primers | 12 places |
| Sealing Film | / | 1 sheet |
| Isothermal Amplification Reagent | Fluorescent dye, Enzyme | 270 µL/tube |
| Positive Control | Escherichia coli Genomic DNA | 160 µL/tube |
Table 2: Composition of nucleic acid detection kit for respiratory tract pathogens.
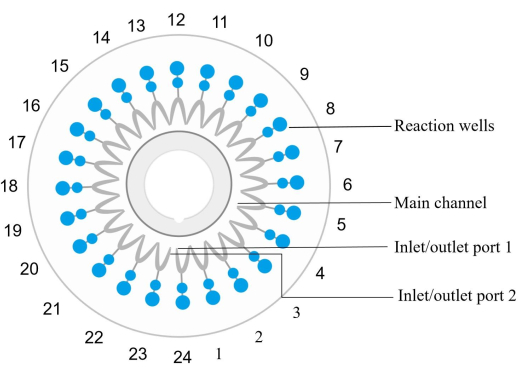
Figure 2: Disc chip structure diagram. The reaction wells are sequentially numbered counterclockwise, where the inlet/outlet port 1 corresponds to the reaction well number 1. Reaction wells 1, 4, 7, 10, 13, 16, 19, 22, and 24 are negative controls. Reaction well 6 is a positive control (E. coli). The reaction well 12 is an internal positive control, and reaction well 23 is an external positive control. Reaction well 2 detects spn. Reaction well 3 detects sau. The reaction well 5 detects MRSA. The reaction well 8 detects kpn. The reaction well 9 detects pae. The reaction well 11 detects aba. The reaction well 14 detects sma. The reaction well 15 detects hin. Please see Table 5 for sample details. Please click here to view a larger version of this figure.
4. Sample preparation and bacterial detection
- Prepare the isothermal amplification reaction system
- Take the isothermal amplification reagent from the kit and allow it to thaw at room temperature fully. Gently shake to mix thoroughly and centrifuge briefly to collect at the bottom of the tube.
- In the reagent storage and preparation area, pipette 20 µL of the isothermal amplification reagent into a prepared 200 µL centrifuge tube. Cover the tube and move it to the specimen preparation area with one centrifuge tube per sample.
- In the specimen preparation area, add 34.5 µL of the target nucleic acid sample. Shake gently to mix thoroughly and centrifuge briefly to collect at the bottom of the tube. The total volume of each nucleic acid amplification reaction system is 54.5 µL (Table 3).
- Chip loading procedure
- On the chip packaging, label the sample number. Open the microfluidic chip with the packaging label facing up.
- Place the chip with the inlet and outlet ports facing upwards. Using a pipette, draw 50 µL of the prepared amplification reaction system and add it to the main channel of the chip through the inlet port. Stop adding when the main channel is filled, and quickly wipe off any excess liquid around the inlet and outlet ports with lint-free tissue.
- Use tweezers to pick up one sealing film and cover it over the inlet and outlet ports. Press a clean pipette tip onto the sealing film in one direction. Ensure a tight seal.
- qPCR reaction
- After the light source of the nucleic acid analyzer has completed preheating, click on the Open Tray button. Place the chip with the front side up on the tray, ensuring that the locating small cylinder emerges from the center gap of the chip to secure it. Align the center positioning device in the tray with the large central hole in the chip.
- Click on the Close Tray button to insert the chip into the nucleic acid analyzer.
- In the Sample Information area on the detection interface, enter the information for testing the sample. Sample number, chip number, and sample type are mandatory; others are optional.
- Click on the Start Detection button in the operation area to initiate the sample detection. The instrument will conduct sample testing according to the preset program.
NOTE: After the completion of detection, the accompanying software of the device will automatically conduct data analysis. Simultaneously, the nucleic acid analyzer will initiate an automatic cooling process. Once the temperature drops to 37 °C, the instrument automatically opens the tray and ejects the chip for retrieval. The results will be interpreted automatically by the system (Table 4).
- Quality control standards
- One positive quality control and nine negative quality controls are enclosed in each chip in this reagent kit. Ensure that the detection results for the positive quality control is positive, and simultaneously, the results for all nine negative quality controls are negative.
NOTE: The results are displayed on the right side of the image stating "Quality Control Results: Experiment Normal (Positive Normal, Negative Normal)," indicating that the test results are valid. Retest is required if any result is incorrect and the sample test result is deemed invalid. One positive internal control using human-specific primers is enclosed in the chip, the result of which is positive for clinical sample testing while being negative if the sample has a low human genomic DNA content, indicating a lower cell count. Recollect and retest samples in such cases.
- One positive quality control and nine negative quality controls are enclosed in each chip in this reagent kit. Ensure that the detection results for the positive quality control is positive, and simultaneously, the results for all nine negative quality controls are negative.
- Results analysis
- After completion of the detection, utilize the second-derivative maximum method combined with commercially protected algorithms in the software to calculate the first inflection point of the S-shaped amplification curve during the rapid amplification phase. Details are described in the "Principles of the Test" part of Respiratory Pathogens Nucleic Acid Detection Kit Instructions (Supplementary File 1).
NOTE: The TP value is the time difference between the inflection point and the origin. The result is interpreted based on the TP and positive judgment values. If the TP value for a detection index is less than or equal to the positive judgment value for that index, it is interpreted as positive. If the TP value exceeds the positive judgment value, it is interpreted as negative, according to the criteria for "positive decision value" in the instructions. The "Fluorescence Curve Area" displays the normalized curve, and the "Detection Result" area shows the quality control and detection results for each parameter (Table 5).
- After completion of the detection, utilize the second-derivative maximum method combined with commercially protected algorithms in the software to calculate the first inflection point of the S-shaped amplification curve during the rapid amplification phase. Details are described in the "Principles of the Test" part of Respiratory Pathogens Nucleic Acid Detection Kit Instructions (Supplementary File 1).
| Reactant | Volume (µl) |
| Isothermal Amplification Reagent | 20 |
| Template DNA | 34.5 |
Table 3: Isothermal amplification reaction system.
| Step | One | Two |
| Temperature (°C) | 37 | 65 |
| Time (min) | 3 | 47 |
Table 4: Nucleic acid amplification reaction program.
| Indicator Name | Positive Control Value |
| Streptococcus pneumoniae (sp) | 30 |
| Staphylococcus aureus (sau) | 34 |
| Methicillin-resistant Staphylococcus aureus (mrsa) | 22 |
| Klebsiella pneumoniae (kpn) | 29 |
| Pseudomonas aeruginosa (pae) | 36 |
| Acinetobacter baumannii (aba) | 36 |
| Stenotrophomonas maltophilia (sma) | 27 |
| Haemophilus influenzae (hin) | 36 |
Table 5: Positive control value for infection indicator.
Results
This experiment employs isothermal amplification technology, conducting reactions on a microfluidic disc chip. The reaction occurs on a microfluidic chip nucleic acid analyzer, employing a fluorescence dye insertion method. The isothermal reaction is performed at a constant temperature of 65 °C, and real-time fluorescence analysis is carried out simultaneously. Positive samples undergo amplification under the action of polymerase with chain displacement functionality, resulting in an S-shaped amplification curve. Th...
Discussion
Respiratory tract infections are prevalent hospital-associated infections, imposing severe consequences on patients and escalating mortality rates16. Timely and accurately identifying potential pathogens followed by effective antibiotics is the key to successful treatment and improving prognosis, particularly given the limitations inherent in traditional culture methods17. In this study, we used a LAMP-based method to determine single or multiple infections for fast and pre...
Disclosures
The authors declare no conflict of interest.
Acknowledgements
We greatly appreciated the financial support provided by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2022A1515220023) and the Research Foundation for Advanced Talents of Guandong Provincial People's Hospital (Grant No. KY012023293).
Materials
| Name | Company | Catalog Number | Comments |
| Bath Incubator(MK2000-2) | ALLSHENG | Provide a constant temperature environment | |
| Bronchial lavage fluid collector head | TIANPINGHUACHANG | SEDA 20172081375 | Collecting bronchoalveolar lavage fluid |
| Fiberoptic bronchoscope | OLYMPUS | SEDA 20153062703 | A flexible bronchoscope equipped with a fiberoptic light source and camera, to visually examine the airways and structures within the lungs. Assist in collecting bronchoalveolar lavage |
HR1500- B2 B2 | Haier | SEDA 20183541642 | Biosafety cabinet |
| NAOH | MACKLIN | S817977 | Liquefy viscous lower respiratory tract sample |
| Nucleic acid detection kit for respiratory tract pathogens | Capitalbio Technology | SEDA 20173401346 | Testing for bacteria infection |
| Nucleic acid extraction reagent | Capitalbio Technology | SEDA 20160034 | For DNA extraction |
| RTisochip-W | Capitalbio Technology | SEDA 20193220539 | Loop-mediated Isothermal Amplification |
| THERMO ST16R | Thermo Fisher Scientific | SEDA 20180585 | Centrifuge the residual liquid off the wall of the tube. |
| Vortex mixer VM-5005 | JOANLAB | For mixing reagent |
References
- GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: A systematic analysis for the global burden of disease study 2016. Lancet Infect Dis. 18 (11), 1191-1210 (2018).
- Niederman, M. S., Torres, A. Respiratory infections. Eur Respir Rev. 31 (166), 220150 (2022).
- Weiser, J. N., Ferreira, D. M., Paton, J. C. Streptococcus pneumoniae: Transmission, colonization and invasion. Nat Rev Microbiol. 16 (6), 355-367 (2018).
- Watt, J. P., et al. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years_ global estimates. Lancet. 374 (9693), 903-911 (2009).
- Kadioglu, A., Weiser, J. N., Paton, J. C., Andrew, P. W. The role of streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol. 6 (4), 288-301 (2008).
- Popova, G., Boskovska, K., Arnaudova-Danevska, I., Smilevska-Spasova, O., Jakovska, T. Sputum quality assessment regarding sputum culture for diagnosing lower respiratory tract infections in children. Open Access Maced J Med Sci. 7 (12), 1926-1930 (2019).
- Nuyttens, H., Cyncynatus, C., Renaudin, H., Pereyre, S., Identification Bébéar, C. expression and serological evaluation of the recombinant ATP synthase beta subunit of mycoplasma pneumoniae. BMC Microbiol. 10 (1), 216 (2010).
- Noviello, S., Huang, D. B. The basics and the advancements in diagnosis of bacterial lower respiratory tract infections. Diagnostics (Basel). 9 (2), 37 (2019).
- Hanson, K. E., et al. Molecular testing for acute respiratory tract infections: Clinical and diagnostic recommendations from the IDSA's diagnostics committee. Clin Infect Dis. 71 (10), 2744-2751 (2020).
- Daniel Reynolds, J. P. B., et al. The threat of multidrug-resistant/extensively drug-resistant gram-negative respiratory infections: Another pandemic. Eur Respir Rev. 31 (166), 220068 (2022).
- T Notomi, H. O., et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 (12), 63 (2000).
- Soroka, M., Wasowicz, B., Rymaszewska, A. Loop-mediated isothermal amplification (LAMP): The better sibling of PCR. Cells. 10 (8), 1931 (2021).
- Mori, Y., Kitao, M., Tomita, N., Notomi, T. Real-time turbidimetry of lamp reaction for quantifying template DNA. Biophys J. 59 (2), 145-157 (2004).
- Parida, M., Sannarangaiah, S., Dash, P. K., Rao, P. V., Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique: Perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 18 (6), 407-421 (2008).
- Chinese Medical Association Respiratory Branch Critical Care Medicine Group, Working Committee on Critical Care Medicine of the Chinese Physicians Association Respiratory Physicians Branch. Standardization of collection, submission, testing, and interpretation of bronchoalveolar lavage fluid in ICU patients. Chin J Tubere Respir Dis. 43 (9), 744-756 (2020).
- Koch, A. M., Nilsen, R. M., Eriksen, H. M., Cox, R. J., Harthug, S. Mortality related to hospital-associated infections in a tertiary hospital: Repeated cross-sectional studies between 2004-2011. Antimicrob Resist Infect Control. 4 (1), 57 (2015).
- Seymour, C. W. F. G., et al. Time to treatment and mortality during mandated emergency care for sepsis. N Engl J Med. 376 (23), 2235-2244 (2017).
- Choi, C. W., Hyun, J. W., Hwang, R. Y., Powell, C. A. Loop-mediated isothermal amplification assay for detection of candidatus Liberibacter asiaticus: A causal agent of citrus huanglongbing. Plant Pathol. 34 (6), 499-505 (2018).
- Büscher, P., Njiru, Z. K. Loop-mediated isothermal amplification technology: Towards point of care diagnostics. PLoS Negl Trop Dis. 6 (6), e1572 (2012).
- Notomi, T., Mori, Y., Tomita, N., Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J Microbiol. 53 (1), 1-5 (2015).
- Ajibola, O., Gulumbe, B., Eze, A., Obishakin, E. Tools for detection of schistosomiasis in resource limited settings. Medical Sciences. 6 (2), 39 (2018).
- Kumar, Y. S. B., et al. Loop-mediated isothermal amplification (LAMP): A rapid and sensitive tool for quality assessment of meat products. Compr Rev Food Sci Food SAF. 16 (6), 1359-1378 (2017).
- Hongling Ou, Y. W., et al. Rapid detection of multiple pathogens by the combined loop-mediated isothermal amplification technology and microfluidic chip technology. Ann Palliat Med. 10 (10), 11053-11066 (2021).
- Liang Wang, J. -. X. L., et al. Quantitative polymerase chain reaction (qPCR)-based rapid diagnosis of helicobacter pylori infection and antibiotic resistance. J Vis Exp. (197), e65689 (2023).
- Jing-Wen Lyu, B. X., et al. Rapid prediction of multidrug-resistant klebsiella pneumoniae through deep learning analysis of SERS spectra. Microbiol Spectr. 11 (2), e0412622 (2023).
- Wei Liu, J. -. W. T., et al. Discrimination between carbapenem-resistant and carbapenem-sensitive klebsiella pneumoniae strains through computational analysis of surface-enhanced Raman spectra a pilot study. Microbiol Spectr. 10 (1), e0240921 (2022).
- Zhang, L. Y., et al. Classification and prediction of klebsiella pneumoniae strains with different MLST allelic profiles via SERS spectral analysis. Peer J. 11, e16161 (2023).
- Zhi Liu, Q. Z., et al. Rapid and sensitive detection of salmonella in chickens using loop-mediated isothermal amplification combined with a lateral flow dipstick. J Microbiol Biotechnol. 29 (3), 454-464 (2019).
- Chen, F., et al. Fully Integrated microfluidic platform for multiplexed detection of hunov by a dynamic confined-space-implemented one-pot RPA-LAMP system. Adv Sci (Weinh). 21, e2306612 (2023).
Reprints and Permissions
Request permission to reuse the text or figures of this JoVE article
Request PermissionThis article has been published
Video Coming Soon
Copyright © 2025 MyJoVE Corporation. All rights reserved